Abstract
The yeast strain SJP-SNU was investigated as a probiotic and was characterized with respect to growth temperature, bile salt resistance, hydrogen sulfide reducing activity, intestinal survival ability and chicken embryo pathogenicity. In addition, we determined the complete genomic and mitochondrial sequences of SJP-SNU and conducted comparative genomics analyses. SJP-SNU grew rapidly at 37 °C and formed colonies on MacConkey agar containing bile salt. SJP-SNU reduced hydrogen sulfide produced by Salmonella serotype Enteritidis and, after being fed to 4-week-old chickens, could be isolated from cecal feces. SJP-SNU did not cause mortality in 10-day-old chicken embryos. From 13 initial contigs, 11 were finally assembled and represented 10 chromosomal sequences and 1 mitochondrial DNA sequence. Comparative genomic analyses revealed that SJP-SNU was a strain of Pichia kudriavzevii. Although SJP-SNU possesses pathogenicity-related genes, they showed very low amino acid sequence identities to those of Candida albicans. Furthermore, SJP-SNU possessed useful genes, such as phytases and cellulase. Thus, SJP-SNU is a useful yeast possessing the basic traits of a probiotic, and further studies to demonstrate its efficacy as a probiotic in the future may be warranted.
Similar content being viewed by others
Introduction
Many yeasts are present in fermented materials, feces and various environmental sources, a number of which have been used in the production of fermented foods, wine and biofuels. Pichia pastoris has been attempted to be used for single cell protein as animal feed additive, while Saccharomyces cerevisiae has been used as a probiotic for farm animals (Ahmad et al. 2014; Chaucheyras-Durand and Fonty 2001). Saccharomyces boulardii has been used as a probiotic in humans and farm animals due to its antibacterial and anti-diarrheal activities (Baum et al. 2002; Kelesidis and Pothoulakis 2012). Compared with S. cerevisiae, S. boulardii grows at body temperature and may be the preferred choice for use as a probiotic in animals. Probiotics were defined as live microorganisms which confer a health benefit on the host and include bacteria, Lactobacillus, Bifidobacterium etc. and yeasts (Ref. FAO/WHO. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Córdoba: FAO/WHO; 2001).
Many fungi have been classified on the basis of morphology, but molecular taxonomy has been used in the classification of yeasts. Recently, comparative genomic studies have unraveled the phylogenetic relationships and evolutionary mechanisms of yeasts. Yeasts have enriched their genetic material by genome duplication, hybridization and the acquisition of foreign genes and diversified their genomes via extensive gene loss and loss of heterozygosity (Butler et al. 2009; Dujon 2010; Gojkovic et al. 2004; Greig et al. 2002; Marinoni et al. 1999; Wolfe and Shields 1997). Yeasts can replicate by sexual and asexual reproduction but have relatively low outcrossing rates (Tsai et al. 2008).
In the present study, we isolated a yeast SJP-SNU from the decomposed leaves of plants and characterized its basic traits for potential use as a probiotic. We also determined the yeast’s complete genome sequence and conducted comparative genomic analyses to understand its phylogenetic and taxonomic relationships with other yeasts and to identify genes related to pathogenicity, metabolism and other useful phenotypic-related genes.
Materials and methods
Yeast, bacteria and media
The SJP-SNU (KCTC 12756BP) strain was isolated from fermented plants and developed for use as a probiotic by Healingbio Co. (14, Jangmun-ro, Yongsan-gu, Seoul, 04393, Korea). The SJP-SNU strain was cultured using YM agar and broth (BD, Sparks, MD, USA) at 37 °C. Saccharomyces cerevisiae (KCTC 7904) and Saccharomyces cerevisiae var. boulardii CNCM I-1079 (S. boulardii, Lallemand Inc., Quebec, Canada) were also cultured using YM agar and broth (BD). Salmonella serotype Enteritidis (SE50), isolated from a chicken, was cultured using MacConkey and TSI agars, and TSA broth (BD). The SJP-SNU strain was deposited to the Korean Collection for Type Cultures (KCTC-12756BP) in Korea.
Comparison of bile salt resistance and anaerobic growth at 25 and 37 °C
For successful settle-down of probiotics in gastrointestinal tract resistance to bile salt is one of necessary requirements (Klaenhammer and Kullen 1999). S. boulardii is known to grow at 37 °C and has been used as a probiotic in humans. To compare the bile salt resistance and growth efficiency at 37 °C of SJP-SNU and S. boulardii, the strains were cultured on MacConkey agar plates containing bile salt at 37 °C, with colony formation being observed for 5 days. To assess the anaerobic growth of SJP-SNU, the strain was streaked onto YM agar plates and incubated at 25 or 37 °C in anaerobic jars with GasPaks for 48 h (BD).
Hydrogen sulfide (H2S) reduction test
H2S is a toxic gas generated by bacteria in the intestine. To test the H2S reduction ability of SJP-SNU, first Salmonella Enteritidis (SE, 4.3 × 109 cfu/ml) was 10-fold serially diluted, and each dilution was inoculated into TSI (Triple sugar iron) agar by stabbing. The minimum bacterial count that generated H2S was determined to be 106 cfu/ml. Next, undiluted cultures of SJP-SNU (8.8 × 109 cfu/ml) and S. boulardii (2.0 × 108 cfu/ml) were 10-fold diluted in PBS. Finally, 450 µl of each yeast dilution and 50 µl of H2S-producing Salmonella serotype Enteritidis (106 cfu/ml) were mixed and inoculated into TSI agar and immediately incubated overnight at 37 °C.
Intestinal viability test
Saccharomyces cerevisiae has been employed as a probiotic for farm animals. To compare the intestinal viability of S. cerevisiae and SJP-SNU, fifteen 4-week-old specific pathogen free (SPF) chickens (Valo, USA) were grouped into three groups (two inoculation and control groups), and orally inoculated with 107 cfu/ml/chicken/day of yeast for 5 days. At 7 days post-inoculation (dpi), cecal feces were collected and diluted by 10-fold with sterilized PBS. Diluted feces were spread onto YM agar plates containing antibiotics (ampicillin 50 µg/ml, tetracycline 50 µg/ml, gentamicin 50 µg/ml, kanamycin 50 µg/ml, and streptomycin 50 µg/ml) and were incubated at 37 °C or 25 °C for growth of SJP-SNU and S. cerevisiae, respectively. The cfu/g of feces was calculated by multiplying the dilution factor and the number of cfu. Animal protocols used in the study were approved by the Biopoa Co., Ltd. institutional IACUC and performed in accordance with all relevant policies.
Chicken embryo pathogenicity test
To evaluate the pathogenicity of yeast or fungi chicken embryos were inoculated onto chorioallantoic membrane (CAM) but we selected more aggressive route into allantoic cavity (Alexander 1989; Jacobsen et al. 2010). Six 10-day-old SPF embryonated chicken eggs (ECEs, Charles River Laboratories, North Franklin, USA) were inoculated with 105 cfu/egg of SJP-SNU or PBS via the allantoic cavity and were candled twice a day for 60 h during incubation at 37 °C to assess embryo survival. After 3 days of incubation, ECEs were chilled at 4 °C overnight and embryos lesions were observed.
Genome sequencing and assembly
Total DNA of an overnight culture of SJP-SNU was extracted using a DNeasy® Blood & tissue kit (QIAGEN), and PacBio RS II single molecule real time (SMRT) sequencing of SJP-SNU was performed (Theragen ETS, Seongnam, Korea). Briefly, 10 μg of yeast genomic DNA was sheared with a Covaris® g-TUBE® device and size-selection for 15–50 kb was performed with a BluePippin system (0.75% DF Marker S1 high-pass 15–20 kb), both done according to the manufacturer’s protocols. SMRTbell template libraries were subsequently prepared using the commercial Template Preparation Kit from Pacific Biosciences Inc. and involved the sequential steps of DNA end repair, adapter ligation and exonuclease digestion of incompletely ligated products. Next, 0.83 nM of the libraries were later annealed to the sequencing primers followed by binding to 50 nM of P4 DNA polymerase, provided in the Template Binding Kit from Pacific Biosciences Inc. For enhanced loading efficiency, 15 pM of the bound complexes were immobilized onto Magbeads (Pacific Biosciences Inc.) prior to loading into the sequencing zero-mode waveguides (ZMWs). The duration for the sequence collection was set at 180 min using the stage start option. Reads with a length of less than 50 bp were filtered out upon acquisition of the sequencing data, and the minimum polymerase read quality was set at 0.75. The SMRT sequencing data were assembled de novo using the FALCON and HGAP3 software pipelines, and the results were merged and reconciled with GARM metaassembler.
Generation and sequencing of a HiSeq DNAPCR-free library
Each sequenced sample was prepared according to the Illumina protocols. The quantification of DNA and the DNA quality was measured by PicoGreen and Nanodrop. Briefly, one microgram of genomic DNA was fragmented by a Covaris device to obtain 350 bp-sized fragments. The fragmented DNA was blunt-ended and phosphorylated, followed by end repair, and the appropriate library size was selected using different ratios of the sample purification beads. A single ‘A’ was ligated to the 3′ ends of DNA fragments, then Illumina adapters were ligated to the fragments. The final ligated product was then quantified using qPCR according to the qPCR Quantification Protocol Guide and was validated using an Agilent Technologies 2100 Bioanalyzer. (Agilent Technologies, Palo Alto CA, USA). Sequencing was performed using a HiSeq™ 2000 platform (Illumina, San Diego, USA).
The genome sequence data obtained from SMRT sequencing were used for all genome analyses performed in the present study. However, only data obtained from the Illumina HiSeq II was used for pathogenic gene analyses due to its high sequence fidelity.
Prediction of repeats and non-coding RNAs (ncRNAs)
For the repeat composition analysis, we used reference based (RepeatMasker ver. 4.0.7 and RepBase (14,031) library; Institute for System Biology) and de novo based (RepeatModeler, ver. 1.0.8; Institute for System Biology) methods, and the results were combined. Simple sequence repeats (SSRs) were searched in the sequenced genome of SJP-SNU by using SSR Finder (minimal number of repeats was 5). The tRNA sequences were predicted by comparing nucleotide homology with tRNAscan-SE2.0 (http://lowelab.ucsc.edu/tRNAscan-SE/). Small nuclear RNA (snRNA) structure and sequence similarities were assessed using the INFERNAL Tool and rfam database (http://eddylab.org/infernal/). Ribosomal RNA (rRNA) sequences were searched by comparing homology with BLAST.
Prediction and annotation of gene structure
The proteins in the genomes of related yeasts identified by taxonomy profiling were extracted from the NCBI non-redundant (nr) protein database and genes were predicted using the exonerate tool. The final gene set was established by combining cording partial information of combining exonerate tool and intron information of protein hint with gene model of AUGUSTUS (Stanke et al. 2004). Gene annotation was performed by homology search of gene set against UniProt, NCBI nr and InterProScan databases.
In silico characterization of pathogenic and biologically important genes
The pathogenicity-related genes of Candida albicans were selected and nucleotide sequences were collected from GenBank databases (Navarro-Garcia et al. 2001). Nucleotide sequences of collected genes were translated with the program BioEdit. By searching for the pathogenic and biologically important genes and protein names in the gene annotation data of SJP-SNU, homologous genes were collected to compare amino acid sequences using the BLASTP search program.
Results
Comparison of growth temperature, anaerobic growth and bile salt resistance
The growth of SJP-SNU at 37 °C was compared with S. boulardii. SJP-SNU formed visible large colonies after 18 h of incubation while S. boulardii formed small colonies only after 32 h incubation. Therefore, the growth rate of SJP-SNU was greater than S. boulardii at 37 °C (Fig. 1a).
Growth characteristics of SJP-SNU. a Comparison of growth efficiency of SJP-SNU and S. boulardii at 37 °C on YM agar plates (1: SJP-SNU after 18 h, 2: S. boulardii after 18 h., 3: S. boulardii after 32 h.). b Anaerobic growth of SJP-SNU on YM agar plates (1: Aerobic condition at 37 °C for 48 h., 2: Anaerobic condition at 37 °C for 48 h.). c Comparison of bile salt resistance of SJP-SNU and S. boulardii cultured at 37 °C on MacConkey agar plates for 5 days (1: SJP-SNU, 2: S. boulardii)
Because S. cerevisiae is known to grow under anaerobic conditions, the anaerobic growth of SJP-SNU was compared with S. cerevisiae. SJP-SNU and S. cerevisiae were incubated anaerobically at 37 and 25 °C, respectively, for 48 h on YM plates. SJP-SNU formed large visible colonies under both aerobic and anaerobic conditions (Fig. 1b).
SJP-SNU and S. boulardii were cultured on MacConkey agar plates at 37 °C for 5 days. While SJP-SNU formed visible colonies, S. boulardii did not. Thus, the growth of SJP-SNU was retarded but not completely inhibited (Fig. 1c).
Hydrogen sulfide (H2S) reduction ability of SJP-SNU strain
TSI medium contains ferrous sulfate as an indicator of H2S production by inoculated bacteria. SJP-SNU and S. boulardii were tested for H2S-reducing activity. Co-cultures of either SJP-SNU or S. boulardii with H2S-producing Salmonella Enteritidis decreased the blackish discoloration of the TSI media. The 100-fold- (8.8 × 107) and 10-fold-diluted (2.0 × 107) SJP and S. boulardii, respectively, suppressed completely the discoloration of media (Fig. 2).
H2S removal activity of SJP-SNU and S. boulardii. SJP-SNU (8.8 × 109 cfu/ml, a) and S. boulardii (2 × 108 cfu/ml, b) were diluted 10-fold (1: 1, 2: 10−1, 3: 10−2, 4: 10−3, 5: 10−4) and co-cultured with H2S-prodcing Salmonella serotype Enteritidis (106 cfu/ml) in TSI agar medium at 37 °C for 18 h. The red bar separates H2S-negative (left) and H2S-positive (right) tubes
Comparison of intestinal viability of S. cerevisiae and SJP-SNU in chicken
Probiotic microorganisms need to survive in the intestines to provide positive effects for hosts. We orally administered 107 cfu/ml of SJP-SNU and S. cerevisiae to 4-week-old SPF chickens daily for 5 days and the number of yeast in feces were counted using YM agar plates containing multiple antibiotics. No yeasts were isolated in the cecal feces of the negative control and S. cerevisiae-administered chickens, whereas yeasts were isolated from the cecal feces of SJP-SNU-administered chickens (Table 1).
Pathogenicity of SJP-SNU in chicken embryos
The pathogenicity of SJP-SNU was tested in embryonated chicken eggs (ECEs). The SJP-SNU strain was inoculated via the allantoic cavity route and did not cause mortality of embryos for 3 days and the embryos did not show any congestion, hemorrhaging or body atrophy. Thus, SJP-SNU had no pathogenic effect on chicken embryos.
Structure and functional annotation of the SJP-SNU genome
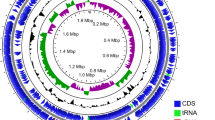
The PacBio sequencing and assembly pipelines generated 13 contigs of 11,005,966 bases (depth of 78.6 and a N50 of 1,367,155 bp) and we conducted sequence comparisons by querying paired contigs in BLATN programs. We observed that contig 13 (22,460 bp) was a part of contig 12 (56,499 bp) and more than the first half (36,004 bp) of contig 12 overlapped the 3′ end of contig 1. We joined the remaining half of contig 12 to contig 1. In addition, we found overlapping sequences at the both ends of contig 11 and removed the repeated sequences at the 3′-end (24,003 bp). The final size (10,923,756 bp) and GC content (38.2%) of the 11 contigs were summarized in Table 2. The genome sequence of SJP-SNU strain was deposited in Genebank under the accession number (SUB2596880; PRJNA383123; SAMN06675446). In total, 3878 gene models (3713 of unique genes and 165 isoforms) were predicted and 3590 genes were annotated. The average gene length was 1320 bp and the total bases of gene models was 5.12 Mbp (46.53% of the draft genome). The number of exons was 6012 (1.55 exon/gene on average) and the average exon length was 716 bp. The number of introns was 2134 (0.55 intron/gene on average) and the average exon length was 380 bp. The exons and introns covered 39.15 and 7.38% of the draft genome, respectively.
Retrotransposons and unclassified and simple repeats were predicted and covered 2.87% of the genome (Additional file 1: Table S1). The copy numbers of ribosomal RNA (rRNA), transfer RNA (tRNA), and small nuclear RNA (snRNA) were 29, 209 and 257, and covered 0.32, 0.14 and 0.19% of genome, respectively (Additional file 1: Table S2). The genes related to the biological characteristics and pathogenicity of SJP-SNU were deposited in Genebank under the accession number (Tables 3 and 4).
Molecular taxonomy of SJP-SNU on the basis of genome sequences
The nucleotide sequences of contigs 1–11 were compared with the genome sequences of yeasts in the GenBank databases. All of the contigs 1–11 showed the highest similarity to P. kudriavzevii (taxID 4909) with high e-values. Therefore, SJP-SNU is a strain of P. kudriavzevii with basic probiotic traits.
Characterization of biologically useful genes
Anaerobic growth of probiotic yeasts is required for growth and metabolism in intestinal environments. To date, genes required only under anaerobic conditions have been reported in S. cerevisiae, and we searched for these genes in the annotated genes of SJP-SNU (Ishtar Snoek and Yde Steensma 2007). We identified homologous genes that are essential for anaerobic growth, but not-essential for aerobic growth, which are summarized in Table 3. Phytase has been used as a feed additive to increase the digestion of inorganic phosphates in feed, and SJP-SNU possessed four phytase homologs in its genome. Cellulase, endo-1,3(4)-beta-glucanase and glucoamylase cleave the glycosidic bonds of cellulose, cereal d-glucans and starch, respectively, to generate glucose and may improve the feed utilization of farm animals (Casey and Walsh 2004). SJP-SNU possessed four cellulase homologs, one endo-1,3(4)-beta-glucanase and one glucoamylase homologs (Table 3). P. kudriavzevii homologs that are required for bioethanol production by xylose fermentation were also identified in SJP-SNU (Chan et al. 2012).
Comparison of pathogenicity-related genes of SJP-SNU to C. albicans
To date, various pathogenicity-related genes have been characterized in C. albicans, and we compared these to the amino acid sequences of SJP-SNU homologs. Pathogenicity-related genes with more than 90% coverage were selected, and the percentiles of coverage and identity and e-values are summarized in Table 4. SJP-SNU possessed pathogenicity-related genes involved in metabolism (FAS2, FET3, FTR1, HEM3, HIS1, LEU2, URA3, TPS1), cell wall synthesis (CHS3, GNA1, MNT1, PHR1, BGL2), signal transduction (TUP1, CEK1, HOG1) and other functions (CTA1, RSR1, PLB1, SAP2, TOP1), although most of the amino acid sequence identities of the SJP-SNU genes were very low (22–80%) (Table 4).
Discussion
According to the comparative genomics study SJP was identified as P. kudriavzevii. P. kudriavzevii has been isolated from fermented foods and fruit juices and is known as Issatchenkia orientalis and is an anamorph of C. krusei (Arias et al. 2002; Carlotti et al. 1996; Chanprasartsuk et al. 2010; Meroth et al. 2003; Mugula et al. 2003; Zott et al. 2010). P. kudriavzevii is heterothallic but is known to be fertile with P. membranaefaciens, P. scutulata, Candida lambica, C. diversa, C. ingens, C. silvae, C. valida, C. vini, C. norvegensis, or Torulopsis inconspicua (Kurtzman and Smiley 1976).
Live S. cerevisiae has been fed to farm animals as a source of vitamins and amino acids and as a probiotic for a long time. S. cerevisiae can replicate in anaerobic conditions, although its optimal temperature for active replication and metabolism is lower than the body temperature of farm animals (37–41 °C). The genome of S. boulardii is more than 99% similar to the genome to S. cerevisiae but it can grow at 37 °C and has been used as a probiotic in humans and farm animals (Kelesidis and Pothoulakis 2012). In the present study, we compared the growth characteristics of SJP-SNU to S. cerevisiae and S. boulardii. SJP-SNU grew more rapidly than S. boulardii on YM and MacConkey agar plates at 37 °C, and both SJP-SNU and S. cerevisiae grew in anaerobic conditions. Thus, SJP-SNU possessed the basic essential traits for probiotics usage. These basic essential traits were demonstrated by the re-isolation of a high number of SJP-SNU isolates from cecal feces. In comparison, S. cerevisiae was not isolated from cecal feces. To date, various S. cerevisiae genes related to anaerobiosis have been reported. Cytoplasmatic dihydroorotate dehydrogenase (DHODase, URA1), involved in de novo pyrimidine biosynthesis, is related to the oxygen-independent growth of S. cerevisiae (Gojkovic et al. 2004). Furthermore, dozens of genes essential for anaerobic growth that are not essential for aerobic growth have been reported (Ishtar Snoek and Yde Steensma 2007). Rapid anaerobic growth of yeasts is not common, but S. cerevisiae can grow rapidly in anaerobic conditions. SJP-SNU possessed no URA1 but possessed homologs of essential genes for anaerobiosis (Table 3). Therefore, these traits may be related to the presence of SJP-SNU at high numbers in cecal feces.
Ethanol has been used as an antimicrobial agent in foods and is known to inhibit Listeria monocytogenes (Oh and Marshall 1993). Yeasts generate ethanol from glucose by alcoholic fermentation under anaerobic conditions. SJP-SNU possess genes involved in alcoholic fermentation using glucose and additional genes (xylose reductase and xylulose kinase genes) for ethanol production with xylose. Therefore, SJP-SNU may produce ethanol in the anaerobic intestines, which may affect microbiota of intestines.
Hydrogen sulfide (H2S) is an irritant gas generated under anaerobic conditions by microorganisms in intestines and feces and is a by-product of alcoholic fermentation of yeast. H2S is cytotoxic that depletes glutathione, an antioxidant, and increases intracellular iron and reactive oxygen species (Truong et al. 2006). The amount of H2S produced by S. cerevisiae depends on the strain and nutrient availability, and MET17 plays a role in the conversion of sulfide to cysteine (Cherest and Surdin-Kerjan 1992; Wainwright 1971). SJP-SNU and S. boulardii possessed MET17 in their genomes, and their H2S reducing activities may be related to MET17. Therefore, the H2S reducing activity of SJP-SNU may be valuable trait as a probiotic.
The addition of exogenous cellulase, beta-glucanase and glucoamylase improved digestibility and utilization of feed for farm animals (Kuhad et al. 2011; Mathlouthi et al. 2003; Rojo et al. 2005). SJP-SNU possesses cellulase, beta-glucanase and glucoamylase, and various glucans in ingested feed may be digested efficiently. Phytases catalyze the hydrolysis of phytic acid in feed grain and improve the utilization of digested inorganic phosphorus by farm animals. Therefore, studies on the bioactivities of SJP-SNU phytases and their effect on farm animal productivity may be valuable in the future.
Candida albicans is pathogenic, and other opportunistic pathogenic yeasts have been reported. To date, several categories of pathogenicity-related genes of C. albicans have been reported (Navarro-Garcia et al. 2001). We collected the homologs of the C. albicans pathogenicity-related genes from gene annotation data of SJP-SNU and compared their amino acid sequences. The genes with more than 90% coverage and e-values are summarized in Table 4. Although the e-values are sufficiently high to support that they are homologs of C. albicans genes, the amino acid identities are not sufficiently high to extrapolate that they have the same pathogenic roles. SJP-SNU possessed the most similar pathogenicity-related genes to those of P. kudriavzevii, but the frequency of P. kudriavzevii in clinical cases is less than C. albicans. A major concern of P. kudriavzevii (C. krusei) infections is multi-drug resistance, and the L656C mutation of a glucan synthase (FSK1) is related to resistance to echinocandin (Kahn et al. 2007). SJP-SNU has same FSK1 gene as P. kudriavzevii, but does not have the L656C mutation. Although we can not make any conclusions on the opportunistic pathogenicity of SJP-SNU, the lack of mortality and pathogenic lesions of 10-day-old chicken embryos may reflect a low pathogenicity of SJP-SNU. In addition, how the chimeric chromosome composition affects the pathogenicity of SJP-SNU may be the subject future study.
Thus, SJP-SNU is a novel yeast that possesses the basic traits of a probiotic, and the genome data obtained in this study may be useful for understanding the evolution and genotype–phenotype correlation of yeasts.
References
Ahmad M, Hirz M, Pichler H, Schwab H (2014) Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol 98(12):5301–5317
Alexander DJ (1989) Newcastle disease. American Association of Avian Pathologists, Prince Albert
Arias CR, Burns JK, Friedrich LM, Goodrich RM, Parish ME (2002) Yeast species associated with orange juice: evaluation of different identification methods. Appl Environ Microbiol 68(4):1955–1961
Baum B, Liebler-Tenorio EM, Enss ML, Pohlenz JF, Breves G (2002) Saccharomyces boulardii and bacillus cereus var. Toyoi influence the morphology and the mucins of the intestine of pigs. Z Gastroenterol 40(5):277–284
Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA (2009) Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459(7247):657–662
Carlotti A, Couble A, Domingo J, Miroy K, Villard J (1996) Species-specific identification of Candida krusei by hybridization with the CkF1,2 DNA probe. J Clin Microbiol 34(7):1726–1731
Casey A, Walsh G (2004) Identification and characterization of a phytase of potential commercial interest. J Biotechnol 110(3):313–322
Chan GF, Gan HM, Ling HL, Rashid NA (2012) Genome sequence of Pichia kudriavzevii M12, a potential producer of bioethanol and phytase. Eukaryot Cell 11(10):1300–1301
Chanprasartsuk OO, Prakitchaiwattana C, Sanguandeekul R, Fleet GH (2010) Autochthonous yeasts associated with mature pineapple fruits, freshly crushed juice and their ferments; and the chemical changes during natural fermentation. Bioresour Technol 101(19):7500–7509
Chaucheyras-Durand F, Fonty G (2001) Establishment of cellulolytic bacteria and development of fermentative activities in the rumen of gnotobiotically-reared lambs receiving the microbial additive Saccharomyces cerevisiae CNCM I-1077. Reprod Nutr Dev 41(1):57–68
Cherest H, Surdin-Kerjan Y (1992) Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating of the sulfur metabolism pathway. Genetics 130(1):51–58
Dujon B (2010) Yeast evolutionary genomics. Nat Rev Genet 11(7):512–524
Gojkovic Z, Knecht W, Zameitat E, Warneboldt J, Coutelis JB, Pynyaha Y, Neuveglise C, Moller K, Loffler M, Piskur J (2004) Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Mol Genet Genomics 271(4):387–393
Greig D, Louis EJ, Borts RH, Travisano M (2002) Hybrid speciation in experimental populations of yeast. Science 298(5599):1773–1775
Ishtar Snoek IS, Yde Steensma H (2007) Factors involved in anaerobic growth of Saccharomyces cerevisiae. Yeast 24(1):1–10
Jacobsen ID, Grosse K, Slesiona S, Hube B, Berndt A, Brock M (2010) Embryonated eggs as an alternative infection model to investigate Aspergillus fumigatus virulence. Infect Immun 78(7):2995–3006
Kahn JN, Garcia-Effron G, Hsu MJ, Park S, Marr KA, Perlin DS (2007) Acquired echinocandin resistance in a Candida krusei isolate due to modification of glucan synthase. Antimicrob Agents Chemother 51(5):1876–1878
Kelesidis T, Pothoulakis C (2012) Efficacy and safety of the probiotic Saccharomyces boulardii for the prevention and therapy of gastrointestinal disorders. Therap Adv Gastroenterol 5(2):111–125
Klaenhammer TR, Kullen MJ (1999) Selection and design of probiotics. Int J Food Microbiol 50(1–2):45–57
Kuhad RC, Gupta R, Singh A (2011) Microbial cellulases and their industrial applications. Enzyme Res 2011:280696
Kurtzman CP, Smiley MJ (1976) Heterothallism in Pichia kudriavzevii and Pichia terricola. Antonie Van Leeuwenhoek 42(3):355–363
Marinoni G, Manuel M, Petersen RF, Hvidtfeldt J, Sulo P, Piskur J (1999) Horizontal transfer of genetic material among Saccharomyces yeasts. J Bacteriol 181(20):6488–6496
Mathlouthi N, Juin H, Larbier M (2003) Effect of xylanase and beta-glucanase supplementation of wheat- or wheat- and barley-based diets on the performance of male turkeys. Br Poult Sci 44(2):291–298
Meroth CB, Hammes WP, Hertel C (2003) Identification and population dynamics of yeasts in sourdough fermentation processes by PCR-denaturing gradient gel electrophoresis. Appl Environ Microbiol 69(12):7453–7461
Mugula JK, Narvhus JA, Sorhaug T (2003) Use of starter cultures of lactic acid bacteria and yeasts in the preparation of togwa, a Tanzanian fermented food. Int J Food Microbiol 83(3):307–318
Navarro-Garcia F, Sanchez M, Nombela C, Pla J (2001) Virulence genes in the pathogenic yeast Candida albicans. FEMS Microbiol Rev 25(2):245–268
Oh DH, Marshall DL (1993) Antimicrobial activity of ethanol, glycerol monolaurate or lactic acid against Listeria monocytogenes. Int J Food Microbiol 20(4):239–246
Rojo R, Mendoza GD, Gonz’alez SS, Landois L, B’arcena R, Crosby MM (2005) Effects of exogenous amylases from Bacillus licheniformis and Aspergillus niger on ruminal starch digestion and lamb performance. Anim Feed Sci Technol 123–124:655–665
Stanke M, Steinkamp R, Waack S, Morgenstern B (2004) AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res 32(Web Server issue):W309–W312
Truong DH, Eghbal MA, Hindmarsh W, Roth SH, O’Brien PJ (2006) Molecular mechanisms of hydrogen sulfide toxicity. Drug Metab Rev 38(4):733–744
Tsai IJ, Bensasson D, Burt A, Koufopanou V (2008) Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proc Natl Acad Sci USA 105(12):4957–4962
Wainwright T (1971) Symposium on microbial changes in foods. Production of H2S by yeasts: role of nutrients. J Appl Bacteriol 34(1):161–171
Wolfe KH, Shields DC (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387(6634):708–713
Zott K, Claisse O, Lucas P, Coulon J, Lonvaud-Funel A, Masneuf-Pomarede I (2010) Characterization of the yeast ecosystem in grape must and wine using real-time PCR. Food Microbiol 27(5):559–567
Authors’ contributions
S-MH, W-JS, and IK carried out the experiments; S-MH, H-JK, and J-HK wrote the paper; S-MH, H-JK, and J-HK designed the research; J-HK, H-JK, and S-JP obtained funding. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by a grant (Grant No. 112102-2) from the Ministry of Food, Agriculture, Forestry, and Fisheries, Republic of Korea and Seoul National University (BK21).
Competing interests
There authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Consent for publication
This article does not contain any individual person’s data.
Ethics approval and consent to participate
No human participants were involved in the study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
The genome sequence of SJP-SNU.
Additional file 1: Table S1.
Prediction of repeating sequences in the SJP-SNU genome. Table S2. Prediction of non-coding RNAs.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hong, SM., Kwon, HJ., Park, SJ. et al. Genomic and probiotic characterization of SJP-SNU strain of Pichia kudriavzevii. AMB Expr 8, 80 (2018). https://doi.org/10.1186/s13568-018-0609-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-018-0609-0