Abstract
Serratia proteamaculans CDBB-1961, a gut symbiont from the roundheaded pine beetle Dendroctonus adjunctus, displayed strong cellulolytic activity on agar-plates with carboxymethyl cellulose (CMC) as carbon source. Automatic genome annotation of S. proteamaculans made possible the identification of a single endoglucanase encoding gene, designated spr cel8A. The predicted protein, named Spr Cel8A shows high similarity (59–94 %) to endo-1,4-β-d-glucanases (EC 3.2.1.4) from the glycoside hydrolase family 8 (GH8). The gene spr cel8A has an ORF of 1113 bp, encoding a 371 amino acid residue protein (41.2 kDa) with a signal peptide of 23 amino acid residues. Expression of the gene spr cel8A in Escherichia coli yields a mature recombinant endoglucanase 39 kDa. Cel8A displayed optimal activity at pH 7.0 and 40 °C, with a specific activity of 0.85 U/mg. The enzyme was stable at pH from 4 to 8.5, retaining nearly 40–80 % of its original activity, and exhibited a half-life of 8 days at 40 °C. The K m and V max values for Spr Cel8A were 6.87 mg/ml and 3.5 μmol/min/mg of protein, respectively, using CMC as substrate. The final principle products of Spr Cel8A-mediated hydrolysis of CMC were cellobiose, cello oligosaccharides and a small amount of glucose, suggesting that Spr Cel8A is an endo-β-1,4-glucanase manifesting exo-activity. This is the first report regarding the functional biochemical and molecular characterization of an endoglucanase from S. proteamaculans, found in the gut-associated bacteria community of Dendroctonus bark beetles. These results contribute to improved understanding of the functional role played by this bacterium as a symbiont of bark beetles.
Similar content being viewed by others
Introduction
Dendroctonus bark beetles (Curculionidae: Scolytinae) are natural agents of change in forest ecosystems, as they parasitize and kill damaged, stressed or old pine trees (Fam: Pinaceae), thus promoting natural regeneration of the forest. Some species have great ecological and economic importance, as broadscale outbreaks are common, resulting in extensive mortality of healthy trees. Their life cycle largely takes place within tree bark, where larvae and adults consume phloem, a resource rich in both nutrients and complex molecules including cellulose (15–40 %), hemicellulose and pectin (30–40 %), and lignin (20 %) (Doi and Kosugi 2004).
The digestion of these compounds entails a complex process in which numerous enzymes are involved, consisting of a wide spectrum on specific substrate (Payne et al. 2015). In particular, the cellulose is a linear homopolymer of glucose linked by β-1,4 glycosidic bonds, which generates a zigzag arrangement between neighboring oxygen bridges. The enzymes involved in the depolymerization of cellulose, are mainly endoglucanase (EC 3.2.1.4), exoglucanase (EC 3.2.1.176) (EC 3.2.1.91) and β-glucosidase (EC 3.2.1.21). Endoglucanases hydrolyze glycosidic bonds found in the amorphous regions of the cellulose, generating long chain oligomers for the action of exoglucanases that liberate cellobiose. This is the substrate of β-glucosidase, which results in the production of glucose as the end product (Medie et al. 2012).
Whereas it has been shown that some species of insects from the Isoptera, Coleoptera, Orthoptera and Blattodea orders have the ability to digest cellulose (Sugimura et al. 2003; Kim et al. 2008; Willis et al. 2010; Mei et al. 2016), there is no evidence that bark beetles have this capacity. However, it has been demonstrated that different bacteria from the Cellulomonas, Cellulosimicrobium, Janibacter, and Leifsonia genera, isolated from the adult guts of Dendroctonus valens (Morales-Jiménez et al. 2009), Stenotrophomonas, Ponticoccus, as well as Kocuria isolated from the adult and larval gut of Dendroctonus rhizophagus (Morales-Jiménez et al. 2012), and Serratia, Pseudomonas, Brevundimona, Methylobacterium, and Pseudoxanthomonas isolated from Dendroctonus armandi larval gut (Hu et al. 2014) have cellulolytic capacities.
Bacterial cellulases have attracted much attention because of their varied applicability in industrial processes, including food and brewery production, animal food processing, detergent production and laundry, textile processing and paper pulp manufacture (Juturu and Wu 2014). Likewise, because of the crisis concerning the sustainable supply of fossil fuel and the increased demand for production of biofuels and chemicals from renewable resources, their applications in cellulose biorefinery for producing fermentable sugars are expected to rapidly increase in the foreseeable future (Juturu and Wu 2014; Bhat 2000).
Serratia proteamaculans CDBB-1961, a gram-negative bacteria, isolated from the adult gut of the roundheaded pine beetle, D. adjunctus Blandford, shows outstanding cellulolytic activity in vitro, in comparison to many other bacteria isolated from the guts of other bark beetles (our unpublished results). This activity places S. proteamaculans CDBB-1961 as an excellent study model for testing the hydrolytic capacity of its cellulolytic activity. However, whereas many endoglucanases from different Serratia spp. have been annotated and deposited in the GenBank (NCBI), to our knowledge there are no studies on the molecular and biochemical characterization of these enzymes. Thus, the present study aims to provide a functional characterization of a novel endoglucanase gene from S. proteamaculans in Escherichia coli.
Materials and methods
Chemicals
Culture media were obtained from BD Difco (Sparks, MD, USA) and JT Baker (Phillipsburg, NJ, USA). All other chemicals, solvents and substrates used were of analytical grade and purchased from Sigma-Aldrich (St. Louis, MO, USA) and JT Baker. Kits for expression, DNA isolation and purification, as well as polymerase enzyme and Ni–NTA resin were obtained from Qiagen (Valencia, CA, USA). Restriction enzymes and DNA ladder were purchased from New England Biolabs (Beverly, MA, USA). pJET1.2/blunt vector was purchased from Fermentas (St Leon-Rot, Germany). The chemicals, including the protein molecular weight markers used in the sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and agarose gels analysis and the Econo gradient pump system were purchased from BioRad (Hercules, CA, USA). Lysozyme was obtained from Research Organics (Cleveland, OH, USA) and bovine serum albumin (BSA) was purchased from Pierce (Rockford, IL, USA). Finally, for thin-layer chromatography of silica gel was purchased from Merck KGaA (Darmstadt, Germany).
Isolation and characterization of the Serratia proteamaculans strain
Serratia sp. strain DADMA8 was isolated from the gut of D. adjunctus emerged-adults, which were removed with chisel and hammer from naturally infested Pinus hartwegii trees at 3400 m.a.s.l. in the Sierra del Tigre, Jalisco, México (19º35.7′N, 103º36′W). From a set of 30-guts, serial ten-fold dilutions of the sample were added to phosphate-buffered-saline (PBS) solution, and 100 μl of each dilution were spread on tryptic soy agar (TSA). Plates were incubated at 28 °C for 48–72 h. Based on their morphological characteristics, 60 colonies of bacteria were randomly isolated from the plates. Axenic cultures were stored at −70 °C in 50 % glycerol. Serratia sp. strain DADMA8 was taxonomically identified as S. proteamaculans, based on its 16S rRNA sequence (GenBank accession number AJ233434) and whole genome sequence data analysis. S. proteamaculans was deposited in the National Collection of Microbial Strains and Cellular Cultures (Colección Nacional de Cepas Microbianas y Cultivos Celulares) from CINVESTAV-IPN, Mexico (accession number CDBB-1961).
Bacterial strains, plasmids and growth conditions
Serratia proteamaculans was grown on Congo red agar plates (0.37 g L−1 K2HPO4, 0.27 g L−1 MgSO4, 1.88 g L−1 carboxymethyl cellulose (CMC), 0.2 g L−1 Congo red, 5 g L−1 gelatin, 15 g L−1 agar) and incubated at 28 °C for 24–48 h.
Escherichia coli strains DH5α and the plasmid pJET were used for DNA amplification. E. coli strains JM109 and M15 [pREP4], and expression vector pQE30 were used for protein expression, following the manufacturer’s instructions for the QIAexpressionist kit. E. coli strains were grown in agar plates and/or Luria–Bertani (LB) medium containing ampicillin (100 μg/mL) and/or kanamycin (30 μg/mL) where appropriate, and then they were incubated at 37 °C overnight, unless otherwise stated.
Analysis of gene, putatively involved in cellulose degradation
The genome from S. proteamaculans CDBB-1961 was sequenced and analyzed by Rapid Annotation Subsystem Technology (RAST) (Aziz et al. 2008) and HMMER 3.1b1 software (http://hmmer.org/). A cellulase encoding gene, named spr cel8A, was identified by automatic annotation and deposited in the GenBank (accession number KX023906). The nucleotide sequence of the spr cel8A gene was analyzed and compared to sequence databases by using available online tools (http://www.ncbi.nlm.nih.gov/, http://www.expasy.org).
Analysis of full-length cellulase sequences
Physicochemical characteristics, including molecular mass and isoelectric point (pI) of the predicted protein were determined using the ProtParam program (Gasteiger et al. 2005). Crystal structure data for the cellulase BcsZ from E. coli strain K12 (PDB accession code 3QXF) (http://www.rcsborg/) was used as a template in the ESPript program (Robert and Gouet 2014) to predict the secondary structure elements of Spr Cel8A and other cellulases, showing great similarity to this enzyme. The glycosidase hairpin and active sites were manually identified based on information from the sequence template (3QXF). The likely sub-cellular localization of Spr Cel8A was determined using the TargetP software (Emanuelsson et al. 2000). The Glycopp V1.0 program was used to predict potential N- and O-glycosylation sites (Chauhan et al. 2012).
Cloning of spr cel8A cellulose gene
All molecular methods were performed using standard molecular biology techniques (Sambrook et al. 1989). Genomic DNA from S. proteomaculans was isolated following the protocol provided in DNeasy Blood and Tissue kit. The coding region of spr cel8A gene (GenBank accession number KX023906) (approx 1.1 kb) was amplified by PCR in a programmable thermal controller T100™ Thermocycler (BIO-RAD), using the HotStar Hifidelity, and the following pair of primers: forward, FCQE: 5′ATAGGATCC TGC GAG TGG CCG GCC TGG CAA C 3′, and reverse, RCQE: 5′ GCGAAGCTT TTC GGA AGT TAC GCA TTG GCC G 3′ with restriction sites BamHI and HindIII (underlined), respectively.
The reaction condition consisted of an initial 5 min step at 95 °C followed by 35 cycles of 15-s at 94 °C, 60-s at 82 °C and 90-s at 72 °C, with a final extension for 10-min at 72 °C. The amplicon of ≈1.1 kb was visualized on 0.8 % agarose gels stained with 10 μg/mL EtdBr and compared with a 1 kb base pair (bp) DNA ladder. The amplicon was purified with the QIAquick® Gel extraction kit, cloned into pJET1.2/blunt vector, and transformed into chemically competent E. coli DH5α cells, generating the pJET1.2/spr cel8A construct.
Construction of spr cel8A in the expression vector
The pJET1.2/spr cel8A construct was subjected to double digestion with BamHI and HindIII at 37 °C for 4 h, and the spr cel8A gene was directionally subcloned into the BamHI and HindIII restriction sites of the pQE30 expression vector. Recombinant plasmids (pQE30/spr cel8A) were propagated in E. coli JM109 and purified for DNA sequencing analysis. The pQE30/spr cel8A construct was transferred to E. coli M15 [pREP4] cell for protein expression assays.
Expression of Spr Cel8A cellulase in E. coli
The recombinant cellulase was expressed from E. coli M15 [pREP4] cells harboring the pQE30/spr cel8A construct. Overnight E. coli cultures, grown in LB medium, were diluted 100-fold in LB medium and shake-incubated at 37 °C, 180 rpm, to a OD600nm = 0.6 cell density. IPTG was added to reach a 1 mM final concentration and cultures were further incubated at 28 °C, 180 rpm, overnight. Subsequently, cell cultures were harvested by centrifugation (4000×g at 4 °C for 20 min).
Preparation of cellulase crude extract
Cells contained in 1 mL of culture broth were harvested by centrifugation (4000×g at 4 °C for 20 min) and resuspended in Laemmli sample buffer, prior to being analyzed for total protein extract by 10 % SDS–PAGE following the Laemmli method (Laemmli 1970). Soluble protein was prepared from the bacterial pellet, of the remaining culture broth, resuspended in lysis buffer (100 mM NaCl, 2 mM EDTA and 50 mM Tris–HCl, pH 7.5) at 20:1 (w/v) ratio. Cells were lysed with lysozyme from chicken egg white (10 mg mL−1) at 4 °C for 30 min. Cell debris was removed by centrifugation (10,000×g at 4 °C for 15 min) and the supernatant, which was used as a source of crude cellulase was analyzed in order to reveal cellulase activity, as well as its protein profile, using 10 % SDS–PAGE.
Protein electrophoresis
Protein analyses were carried out by 10 % SDS-PAGE using a Mini PROTEAN II system. Proteins in the gel were visualized by Coomassie Brilliant blue R-250. Protein molecular weight (MW) was estimated with reference to broad range molecular weight protein standards. Gels were recorded and analyzed using a gel documentation system (DigiDoc-It Im aging System, UVP). Protein concentration was determined as described by Lowry et al. (1951), using BSA as standard.
Enzyme assay
The cellulolytic activity of Spr Cel8A on CMC was determined from the amount of reducing sugars released during incubation at 40 °C for 30 min. Reducing sugars were quantified using the dinitrosalicylic acid method, with glucose as standard (Miller 1959). A 20 μL of enzyme preparation was added to 1 mL of 50 mM citrate phosphate buffer, pH 7.0, containing 0.5 % (w/v) CMC as the substrate. One unit (U) of enzymatic activity was defined as the amount of enzyme required to produce 1 μmol of glucose per minute under assay conditions. All tests were carried out in triplicate and average values were recorded.
Purification of a six His-tag recombinant enzyme
Spr Cel8A was purified by affinity chromatography from the soluble fraction of bacterial lysate (E. coli M15 [pREP4]/pQE30/spr cel8A), corresponding to cellulose crude extract.
Crude extract was loaded onto a Ni–NTA resin with a packed volume of 3.37 mL, previously equilibrated with buffer A (400 mM NaCl, 20 mM imidazole, 50 mM sodium phosphate buffer, pH 8.0). The C-terminal His6-tagged protein was subsequently eluted by linear gradient of 30–300 mM imidazole, using an Econo gradient pump system, at a constant flow rate of 1 mL min−1 and 1.0 mL fractions were collected. Fractions with cellulolytic activity were pooled, dialyzed against 50 mM citrate phosphate, pH 7.0, and analyzed by 10 % SDS-PAGE. The purified Spr Cel8A cellulase was stored at 4 °C for further study.
Effect of pH on enzyme activity and stability
The effect of pH on the enzymatic activity of Spr Cel8A was evaluated at pH values, ranging from 3 to 10.5, in CMC (0.5 % w/v) substrate prepared in 50 mM of different buffers: citrate phosphate (pH 3–7), phosphates (pH 6–8) and glycin–NaOH (8.5–10.5), and incubated at 40 °C for 30 min. In order to evaluate the effect of pH on the stability of Spr Cel8A, samples of purified enzyme were incubated in a variety of buffers as mentioned previously, at pH values, ranging from 3.0 to 10.5, at 28 °C for 1 h. Subsequently, the remaining cellulolytic activity was measured under standard conditions and compared to untreated enzyme activity.
Effect of temperature on enzyme activity and stability
The effect of temperature on enzymatic activity of purified Spr Cel8A was determined by conducting the assay at different temperatures ranging from 15 to 55 °C, in 0.5 % (w/v) CMC, dissolved in 50 mM citrate phosphate buffer, at a pH of 7.0, for 30 min. In order to evaluate enzyme thermostability, the purified Spr Cel8A was incubated at different temperatures (30, 40 and 50 °C) in 50 mM citrate phosphate buffer, pH 7.0. For determining the half-life of the enzyme, aliquots of the sample were withdrawn at different time intervals and residual enzymatic activity was measured under standard conditions and compared with the untreated enzyme activity.
Substrate specificity and kinetic parameters
In order to evaluate substrate specificity of Spr Cel8A, cellulolytic activity was determined under optimal assay conditions using 0.5 % (w/v): birchwood xylan, oat spelt xylan, beechwood xylan, CMC, Avicel, Solka floc. For determining kinetic parameters K m and V max , the initial reaction rates for Spr Cel8A were studied under optimal conditions for enzyme activity using CMC as substrate, at a concentration ranging from 0.2 to 1.2 mg. The K m and V max values were obtained by means of the nonlinear least squares regression method using the Michaelis and Menten kinetics (http://biomodel.uah.es/metab/enzimas/MM-regresion.htm).
Effect of ions, EDTA and 2-mercaptoethanol (2-ME) on enzyme activity
In order to study the effect of metal ions and other compounds on the enzymatic activity of Spr Cel8A, samples of the purified enzyme were incubated in 0.5 % CMC prepared in 50 mM citrate phosphate buffer, pH 7.0 containing: Ca2+, Co2+, Mn2+, Ni2+, Mg2+, Fe2+, Cu2+, Na1+, Zn2+, Li1+, Hg2+; EDTA or 2-ME, at a final concentration of 1 and 5 mM each. Reaction mixtures were incubated with the different compounds at 28 °C for 60 min. Cellulase activities were assayed under standard conditions, and compared to a control without additions.
Zymogram
Zymogram analysis was carried out as previously described (Schwarz et al. 1987), with some modifications as follows. Briefly, protein samples were separated in a 10 % polyacrylamide gel co-polymerized with 2 % CMC, under denaturing conditions. Protein samples were resuspended in SDS sample buffer without 2-β-mercaptoethanol, and the samples were boiled on waterbath for 5 min. After electrophoresis, the gels were incubated in 50 mM citrate–phosphate buffer, pH 7.0 at 40 °C for 30 min. Cellulase activity was visualized by staining the gel with Congo red (1 mg/mL) for 15 min, while being gently agitated, and then destained in 1 M NaCl for 10 min.
Analysis of the hydrolysis products of recombinant cellulase using thin-layer chromatography
The degradation pattern and products of recombinant cellulase were analyzed using Thin Layer Chromatography (TLC). Briefly, 50 μL of purified Spr Cel8A cellulase was added to 50 μL of 1 % CMC in 50 mM citrate–phosphate buffer, pH 7.0. The reaction mix was incubated at 40 °C; and aliquot samples were taken for different time spans of between 0 and 144 h. A total of 3 μL of each sample and 2 μL of the cellulose-oligomer standard (glucose, cellobiose or cellotetraose) was spotted on silica gel. The hydrolysis products were separated in a solvent system consisting of butanol/ethanol/water (5:3:2 v/v/v). The plate was then sprayed with sulfuric acid (15 % v/v) and heated in a dry oven at 80 °C for 40 min to visualize the cellulose-oligomers.
3D model of Spr Cel8A from S. proteamaculans
The 3D structure model of endoglucanase was generated by submitting the amino acid sequence of Spr Cel8A from S. proteamaculans to the iterative threading assembly refinement (I-TASSER) server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/).
Results
Analysis of the spr cel8A gene
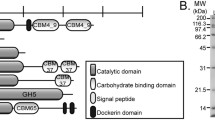
In order to putatively identify the predicted proteins involved in cellulose degradation, the genome of S. proteamaculans (strain DADMA8) was sequenced. Annotation and similarity analysis of the genome sequence allowed the identification of only one gene (spr cel8A), encoding for an endoglucanase precursor (EC 3.2.1.4). The spr cel8A gene from S. proteamaculans has an open reading frame (ORF) of 1113 bp, encoding for a predicted protein of 371 amino acid residues. The molecular mass and theoretical pI of the predicted Spr Cel8A were calculated to be 41.2 kDa and 5.37, respectively, with a signal peptide of 23 amino acids at the N-terminal end and four potential N-glycosylation sites (Fig. 1). Similarity analysis of the amino acid sequence for the predicted protein Spr Cel8A revealed that this enzyme is 93–94 % identical to endoglucanases belonging to the glycosyl hydrolase family 8 (GH8) from S. proteamaculans strain 568 (A8G820, A8G820_SERP5), S. liquefaciens (S5EHX1, S5EHX1_SERLI), S. liquefaciens (A0A109Z435, A0A109Z435_SERLI), and S. grimesii (A0A084YRC6, A0A084YRC6_9ENTR); and 59–61 % identical to GH8 endoglucanases from E. coli O157 (Q8X5L9, GUN_ECO57), E. coli strain K12 (P37651, GUN_ECOLI), S. typhimurium strain L72/SGSC1412/ATCC700720 (Q8ZLB7, GUN_SALTY) and S. typhi (Q8Z289, GUN_SALTI).
Multiple sequence alignment and secondary structure element assignment. The alignment included endoglucanases from Serratia proteamaculans strain 568 (A8G820, A8G820_SERP5), S. liquefaciens (S5EHX1, S5EHX1_SERLI) and E. coli O157 (Q8X5L9, GUN_ECO57). 3D crystal structure of the cellulase BcsZ from Escherichia coli strain K12 (PDB code 3QXF). Assignment of six hairpin glycosidase and active sites were manually determined for comparison with cellulase 3QXF from E. coli strain K12. The α helices are marked as α or β, based on the automatic assignment in conformity with the template of 3QXF protein structure in the ESPript program (Robert and Gouet 2014)
Alignment and comparison of the Spr Cel8A amino acid sequence deduced from S. proteamaculans with that of endoglucanase BcsZ sequence from E. coli strain K12 (P37651, GUN_ECOLI), that has been characterized and crystallized (PDB 3QXF), made possible the identification of 13-α helices, 14-β sheets, six-hairpin glycosidase-like (positions 52–65, 114–132, 179–196, 215–226, 240–249 and 331–354 aa), and a second hairpin (position 114–132), which corresponds to a conserved region in GH8. Amino acid residues E55 and D116 were identified as the proton donor and nucleophile respectively, because of their similarity (Fig. 1).
Expression and purification of a six His-tag recombinant enzyme
The coding region of the spr cel8A gene was expressed from E. coli M15 [pREP-4] cells, harboring the pQE30/spr cel8A construct, for functional analysis of the recombinant protein. The profile of total protein in the soluble fraction of the bacterial lysate revealed the overexpression of polypeptide with an estimated MW of 41 kDa, that corresponds to the expected MW for Spr Cel8A cellulase. In comparison to non-induced cells, the heterologous protein was expressed at high levels by cells, after induction with 1 mM IPTG (Fig. 2a, line 5). The recombinant protein was purified from the soluble fraction of the bacterial crude extract and purified by affinity chromatography on Ni–NTA resin. A summary of the purification steps for Spr Cel8A is presented in Table 1. SDS–PAGE analysis of extracts from the bacteria harboring the construct pQE30/spr cel8A revealed a single protein band of approximately 41 kDa, Spr Cel8A (Fig. 2b). The MW of Spr Cel8A conforms with the theoretical MW of the mature protein (39,008 Da), without the signal peptide. The zymogram analyses of purified Spr Cel8A, performed under denaturing conditions, showed a single band that confirmed the cellulolytic activity and monomeric nature of the enzyme (Fig. 2c).
Protein and zymogram analysis of Spr Cel8A in 10 % SDS-PAGE. a Analysis of total protein extracts of transformed E. coli M15 [pREP4] /pQE30/spr cel8A, M: MW protein standards; line 1 and 2 non-induced soluble fraction medium and bacterial cell harvested respectively; line 3 induced soluble fraction medium; line 4 and 5 induced lysed bacterial cells soluble fraction. b Purification of recombinant Spr Cel8A; line 1 total protein extracts of transformed E. coli M15 [pREP4] /pQE30/Spr Cel8A and line 2 Spr Cel8A purified. c Zimogram analysis of Spr Cel8A in 10 % SDS-PAGE with 2 % CMC
Biochemical characterization of Spr Cel8A protein
The purified Spr Cel8A cellulase from S. proteamaculans expressed in E. coli was biochemically characterized to provide insight concerning its biochemical properties and K m and V max kinetic parameters.
Effect of pH on enzyme activity and stability
Spr Cel8A cellulase displayed optimal activity at pH 7 with a preference for citrate–phosphate buffer; however the enzyme exhibited 40–80 % of its maximal activity at different pH values ranging from 4 to 6.5, and 40 to 60 % at a pH of 7.5–9.5 (Fig. 3a). pH stability assays indicated that Spr Cel8A was highly stable at a pH range between 4 and 8.5, retaining 40–90 % of its original activity after 1 h of incubation at 28 °C (Fig. 3b).
Effect of pH and temperature on Spr Cel8A activity and stability. a pH effect with different buffers; citrate phosphate (pH 3–7), phosphates (pH 6-8) and glycine-NaOH (8.5–10.5) and incubated at 40 ºC for 30 min. b pH stability, protein was incubated in different buffer, mentioned above, at different pH values, ranging from 3.0 to 10.5, at 28 ºC for 1 h. c Temperature effect, the enzyme was incubated with 0.5 % (w/v) CMC dissolved in 50 mM citrate phosphate buffer, pH 7.0, for 30 min at different temperatures (15–55 ºC). All tests were performed in triplicate and error bars indicate standard deviations
Effect of temperature on enzyme activity and stability
Spr Cel8A showed optimal activity at 40 °C (Fig. 3c); however, the enzyme manifested 40–90 % of its original activity at a wide range of temperatures, between 15 and 50 °C (Fig. 3c). The thermal stability of Spr Cel8A was studied at temperature values of 30, 40 and 50 °C at its optimal pH. The enzyme showed a half-life of 8 and 16 days at 40 and 30 °C, respectively (Table 2).
Substrate specificity and kinetic parameters
In order to evaluate the substrate specificity of Spr Cel8A, the activity of the enzyme was assayed using Avicel, CMC, Solka floc, beechwood xylan, birchwood xylan, and oat spelt xylan as the substrate, at a final concentration of 0.5 % each. Spr Cel8A showed higher affinity towards CMC, beechwood xylan and birchwood xylan, while no xylanase activity was detected in the presence of Avicel, Solka floc and oat spelt xylan (Fig. 4a). The kinetic parameters of Spr Cel8A were assayed under optimal pH and temperature conditions with CMC as the substrate, under different concentrations ranging from 0.2 to 1.2 mg/mL. The K m , V max and k cat (V max /[E]) were 6.87 mg/mL, 3.5 μmol/min/mg and 0.089 μmol/min, respectively.
Substrate affinity of Spr Cel8A and effect of metal ions and other agents on enzyme activity. (a) The activity of Spr Cel8A was assayed using Avicel, CMC, Solka Floc, beechwood xylan, birchwood xylan, and oat spelt xylan as the substrate, at a final concentration of 0.5 % each in 50 mM citrate phosphate buffer, pH 7.0 at 40 ºC. (b) Effect of metal ions, EDTA and 2-ME on Spr Cel8A activity at two different concentrations 1 and 5 mM. Enzyme was incubated in 0.5 % (w/v) CMC in 50 mM citrate sodium phosphate buffer, pH 7.0 containing (1 and 5 mM each): Ca2+, Co2+, Mn2+, Ni2+, Mg2+, Fe2+, Cu2+, Na1+, Zn2+, Li1+, and Hg2+; EDTA and 2-ME at 28 ºC for 60 min. All tests were performed in triplicate and error bars indicate standard deviations
Effect of ions, EDTA and 2-mercaptoethanol on enzyme activity
The effect of metal ions, EDTA, and 2-ME, at 1 and 5 mM each, on the activity of Spr Cel8A was evaluated (Fig. 4b). Spr Cel8A activity was enhanced by 108 and 81 % with the ion Co2+ at 1 and 5 mM, respectively; whereas in the presence of the ion Cu2+ (1 mM), the activity of the enzyme increased by 67 %. The metal ions Mn2+ (1 mM), Li1+ (1 and 5 mM), Ca2+ (5 mM), and Na1+ (1 and 5 mM) increased the activity of Spr Cel8A by between 30 and 67 %. The activity of Spr Cel8A was almost completely inhibited in the presence of Zn2+ (1 and 5 mM), Ni2+ (5 mM) and Cu2+ (5 mM) (Fig. 4b); however, the enzyme displayed 64–78 % of its original activity in the presence of Mg2+, Hg2+ and 2-ME, at 1 and 5 mM each. In contrast, the metal ions Ni2+ (1 mM), Mn2+ (5 mM), Ca2+ (1 mM) and Fe2+ (1 and 5 mM), and the quelant agent EDTA (1 and 5 mM) had little or no effect on the activity of Spr Cel8A (Fig. 4b).
Analysis of hydrolysis products by TLC
The hydrolysis products of CMC yielded by the action of Spr Cel8A for different incubation periods were analyzed by TLC. The hydrolysis products released were mainly cellobiose, unknown cello oligosaccharides and a small amount of glucose (Fig. 5).
3D model of Spr Cel8A
Initiating with the Spr Cel8A amino acid sequence from S. proteamaculans, the I-TASSER server generated a 3D structural model of Spr Cel8A with a C-score value of −0.02 (Fig. 6). The 3D model is structurally closest to the endo-1,4-β-d-glucanase from Pseudomonas putida KT2440 (PDB ID: 4Q2B) with a template modeling score (TM-score) of 0.90 and an alignment coverage of 0.90. The 3D model for Spr Cel8A from S. proteamaculans is very rich in α-helices with 50.54 %, followed by random coil structures with 42.71 %, and only about 6.75 % β-sheet content.
Discussion
The metabolic capacities and functional role of gut-associated bacteria from bark beetles have not yet received extensive study, as has been undertaken for other insects such as termites, longhorn beetles, aphids, butterflies and moths (Watanabe and Tokuda 2010; Engel and Moran 2013). The cellulolytic activity of gut-associated bacteria from many insects has been well characterized by both qualitative and quantitative methods (Slaytor 1992; Sun and Scharf 2010); whereas in the case of bark beetles it has been only semiquantitatively determined by CMC degradation (Morales-Jiménez et al. 2009, 2012; Hu et al. 2014; Delalibera et al. 2005). In addition, no research has quantitatively evaluated this activity, or characterized the genes and corresponding enzymes.
In this study, we have identified, purified, characterized, and quantitatively evaluated the activity of an endo-β-1,4-glucanase from S. proteamaculans, isolated from the gut of the bark beetle D. adjunctus. Serratia spp. have been recurrently isolated from the gut of the Dendroctonus species (Morales-Jiménez et al. 2009; Hu et al. 2014; Vasanthakumar et al. 2006; Xu et al. 2015), and their relative abundance and persistence in this habitat suggest that they are part of the microbiome core of these insects (Hernández-García, personal communication). Thus, it appears probable that the cellulolytic activity from Serratia sp. makes a quantitative substantial contribution to the nutrition of these insects.
The complete genome analysis of S. proteamaculans indicates that it contains only a single gene (spr cel8A) involved in cellulose hydrolysis, which has only been observed in 38 % of bacterial genomes deposited in the carbohydrate-active enzyme database (CAZy). For this reason this species was selected for being non saprophytic bacteria that can hydrolyse and synthesize cellulose, as it has an operon which can biosynthesize this polymer, also present in the S. proteamaculans 568 strain (Medie et al. 2012).
The predicted product of the spr cel8A gene, Spr Cel8A, from S. proteamaculans identified in this study shows conserved residues characteristic of the GH8 family (Fig. 1), thus suggesting that Spr Cel8A can be classified as endoglucanase pertaining to the GH8 family. The 3D model of Spr Cel8A from S. proteamaculans (Fig. 6) predicts a 3D structure (α/α)6, closely related to the 3D structure models (α/α)6 of the GH8 endoglucanase family from P. putida KT2440 (UniProtKB accesion code: Q88JL2_PSEPK) and E. coli K-12 (Mazur and Zimmer 2011). Concurring with our results, it has been reported that endoglucanases belonging to the GH8 family present a 3-D structure (α/α)6 (Taylor and Vaisman 2010).
Twenty six endocellulases from the GH8 family have been characterized in terms of bacteria (Berlemont and Martiny 2013), but none of these have been associated with species from the Serratia genus. Therefore, this is the first report of a endo-1,4-β-glucanase from the GH8 family associated with a Serratia species. Similarly, based on molecular and functional characteristics, the endo-1,4-β-glucanase Spr Cel8A from S. proteamaculans should be classified within clan M of the GH8 family, as these are involved in hydrolysis of cellulose by means of a reaction mechanism for inversion (Henrissat 1991; Yennamalli et al. 2011).
Likewise, amino acid identity analyses show that Spr Cel8A endo-1,4-β-glucanase from S. proteamaculans is 94–95 % similar to other endoglucanases annotated from Serratia spp. (Fig. 1), which have not been biochemically characterized. The endo-1,4-β-glucanase Spr Cel8A from S. proteamaculans is 61 % identical to the endoglucanase from E. coli strain K12, whose 3-D structure has been determined by X-ray crystallography (Mazur and Zimmer 2011).
Recombinant endo-1,4-β-glucanase Spr Cel8A from S. proteamaculans displayed remarkable biochemical characteristics that may optimize its performance within the insect’s gut. These include, optimal activity recorded at pH 7.0, cellulolytic activity exhibited over a wide range of pH (4.5–9.5) (Fig. 3a), as well as its high stability within this potential hydrogen interval (Fig. 3b). This suggests that the enzyme may be functional within all gut regions of bark beetles. Our results coincide with that observed in other bacteria such as Komagataeibacter xylinus (Koo et al. 1998) and Bacillus sp. HSH-810 (Kim et al. 2005) isolated from other environments, whose endo-1,4-β-glucanases also manifest optimal activity at pH 7.0 and notable activity at a wide pH ranging from 4.5 to 10. However, these characteristics may apparently constitute an exception, because the rule is that bacterial endo-1,4-β-glucanases work at an optimal pH, but do not show activity at wide intervals of pH, as has been documented in Sporocytophaga myxococcoides (Goksoyr 1988), Pseudomonas fluorescens (Yamane et al. 1970); Ruminiclostridium thermocellum (Romaniec et al. 1992), Aquifex aeolicus (Kim et al. 2000), Ruminococcus albus (Ohara et al. 2000), Cellulomonas biazotea (Rajoka et al. 2004), Clostridium cellulovorans (Arai et al. 2006), Bacillus mycoides (Balasubramanian et al. 2012), and Bacillus sp. (Harshvardhan et al. 2013).
Endoglucanase Spr Cel8A showed pH stability over a wide range of pH from 4 to 8.5; similar results were found among endoglucanases from Bacillus amyloliquefaciens (Lee et al. 2008) and the archaeon from Pyrococcus horikoshii (Yang et al. 2012), with pH stability for a pH range of 4–9.0.
Another interesting biochemical characteristic is the optimal temperature of the β-1,4-endoglucanase Spr Cel8A from S. proteamaculans, with an optimal activity at 40 °C, which maintains 40–90 % of its maximal activity in the temperature range from 15 to 50 °C (Fig. 3c). The optimum temperature recorded for different bacterial endoglucanases shows broad temperature values, ranging from 25 °C in Fibrobacter succinogenes (Lyo and Forsberg 1996) to 100 °C in Thermotoga maritima (Cheng et al. 2012). Findings here indicate that the Spr Cel8A endoglucanase from S. proteamaculans is a mesophilic enzyme.
Thermostability assays indicated that Spr Cel8A displays low thermostability at 50 °C; however the enzyme exhibited half-lifes of 16 and 8 days at 30 and 40 °C, respectively. Findings here are comparable to that reported for the commercial cellulase from Trichoderma reesei, whose activity decreases by 15 % after 150 h in the temperature range from 30 to 50 °C (Balsan et al. 2012). Interestingly, the thermostability of Spr Cel8A at 30 and 40 °C is an attribute that should be emphasized because bark beetle insects spend much of their life cycle under the bark of pine trees feeding on phloem (Wood 1982), where the interior temperature can range between 15 and 40 °C, and their gut temperature varies from 12 to 33 °C (Wermelinger and Seifert 1998). This temperature range conforms to the temperature at which all insect life stages, including egg and adult growth, and development and performance of metabolic processes such as nutrition, take place.
Spr Cel8A was able to hydrolyze CMC and to a lesser extent, beechwood xylan (38 %) and birchwood xylan (2.9 %); however the enzyme was not active on Avicel, Solka floc and oat spelt xylan (Fig. 4a). The K M value of Spr Cel8A was 6.87 mg/mL, which is comparable to that reported for the halophilic cellulase CelB from Bacillus sp. BG-CS10, with a K M value of 6.6 mg/mL, using CMC as substrate with no NaCl (Zhang et al. 2012). However, K M values ranging from 0.01 to 6.6 mg/ml have been reported for endoglucanases, using CMC as the substrate (Zhang et al. 2012; Kupsky et al. 2014).
It is generally accepted that some metal ions and reagents significantly affect cellulase activities (Wang et al. 2012). Therefore, the effect of metal ions, EDTA and 2-mercaptoethanol on the activity of Spr Cel8A was evaluated. Spr CelA from S. proteamaculans was inhibited Zn2+ (1 and 5 mM) or almost completely inhibited by Cu2+ at 5 mM (Fig. 4b). The inhibitory effect of Cu2+ and Zn2+ on the activity of cellulases and xylanases is a common feature among these enzymes (Romaniec et al. 1992; Paradis et al. 1993). However, the activity of Spr Cel8A increased in the presence of Co2+, Mn2+, Li1+ and Na1+ (1 and 5 mM), and other metallic ions such as Cu2+, Ni2+ and Fe2+ at 1 mM, and Ca2+ 5 mM. This suggests that this endoglucanase manifests a conformational stability, because the enzyme retained its activity even in the absence of metallic ions (Welfle et al. 1995; Rubini et al. 2010). An increase of 139.5 % in the activity of an endo-1,4-β-glucanase from B. subtilis was observed in the presence of Co2+ (Au and Chan 1987); whereas Spr CelA from S. proteamaculans increased by 108 %. In Clostridium thermocellum it has also been documented that Ca2+ stabilizes the endoglucanase structural conformation because it has higher affinity for the substrate and great thermostability (Chauvaux et al. 1990). Notably, the enzyme displayed 64–78 % of its original activity in the presence of Mg2+, Hg2+, 2-ME and EDTA at 1 and 5 mM each (Fig. 4b). Enzyme inhibition by metallic cations usually suggests the presence of at least one sulfhydryl group in the active site, usually a cysteine amino acid (Rubini et al. 2010). This group oxidation by the cations destabilizes the conformational folding of the enzyme (Rouvinen et al. 1990) or leads to the formation of disulfide bonds at an irregular position of the protein (Ohmiya et al. 1995). Therefore, the capacity of Spr Cel8A from S. proteamaculans to remain active in the presence of several metal ions and chemical agents, including Hg2+, EDTA and 2-ME, is a fundamental characteristic because many ions are present in the phloem of trees.
Concerning the final hydrolysis products, Spr CelA from S. proteamaculans can be classified as an endo-β-1,4-glucanase that undergoes exo-β-1,4-glucanase activity due to its ability to release cello oligosaccharides, cellobiose and glucose from CMC. Other endo-β-1,4-glucanases that release cello oligosaccharides, cellobiose and glucose from CMC as the main hydrolysis products have been reported previously. For instance, the E4-90 endo-β-1,4-cellulase from Thermomonospora fusca releases mainly cellobiose, cellotriose, cellotetraose and glucose with CMC as the substrate, suggesting that this enzyme is involved in endo- and exo-activity (Irwin et al., 1998), and the endoglucanase Cel9A from Thermobifida fusca releases mainly cellobiose, with some minor products e.g., cellopentaose, cellotetraose, cellotriose with CMC as the substrate (Chir et al. 2011). In contrast, the endocellulase Cel5A from Cellvibrio mixtus releases mainly cellobiose and cellotriose with no evidence of glucose formation during the early stage (Voget et al. 2006).
In summary, we have successfully expressed the endoglucanase Spr Cel8A from S. proteamaculans in E. coli. This enzyme is neutral, mesophilic, displays high affinity to CMC polysaccharide and exhibits exo-activity. Interestingly, Spr Cel8A is highly active in a wide range of pH values (4–9.5) and temperatures (15–50 °C), and has higher thermostability than other cellulases previously reported, such as the commercial cellulose from T. reesei. Additionally, Spr Cel8A is stable in the presence of several metal ions, including Hg2+. This characteristics made this enzyme suitable for structure–function studies. Although many different bacteria endoglucanases have been identified and cloned, only a few are from the genus Serratia, and this is the first report of isolation, cloning and expression of an endo-β-1,4-glucanase from S. proteamaculans, pertaining to the gut-associated bacteria community in Dendroctonus bark beetles. The identification and characterization of this enzyme may improve understanding of the functional role of S. proteamaculans as symbiont of bark beetles.
Abbreviations
- SDS-PAGE:
-
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- BSA:
-
bovine serum albumin
- PBS:
-
phosphate-buffered-saline
- TSA:
-
tryptic soy agar
- CMC:
-
carboxymethyl cellulose
- LB:
-
Luria–Bertani
- RAST:
-
Rapid Annotation Subsystem Technology
- bp:
-
base pair
- IPTG:
-
Isopropyl-β-d-thiogalactopyranoside
- MW:
-
molecular weight
- pI:
-
isoelectric point
- U:
-
unit
- EDTA:
-
ethylenediaminetetraacetic acid
- 2-ME:
-
2-mercaptoethanol
- TLC:
-
Thin Layer Chromatography
- GH8:
-
glycosyl hydrolase family 8
- CAZy:
-
carbohydrate-active enzyme database
References
Arai T, Kosugi A, Chan H, Koukiekolo R, Yukawa H, Inui M. RH properties of cellulosomal family 9 cellulases from Clostridium cellulovorans. Appl Microbiol Biotechnol. 2006;71:654–60. doi:10.1007/s00253-005-0249-6.
Au KS, Chan KY. Purification and properties of the endo-1,4-β-glucanase from Bacillus subtilis. J Gen Microbiol. 1987;133:2155–62.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi:10.1186/1471-2164-9-75.
Balasubramanian N, Toubarro D, Teixeira M, Simos N. Purification and biochemical characterization of a novel thermo-stable carboxymethyl cellulase from Azorean isolate Bacillus mycoides S122C. Appl Biochem Biotechnol. 2012;168:2191–204. doi:10.1007/s12010-012-9929-8.
Balsan G, Astolfi V, Benazzi T, Meireles MA, Maugeri F, Di Luccio M, Dal Prá V, Mossi AJ, Treichel H, Mazutti MA. Characterization of a commercial cellulase for hydrolysis of agroindustrial substrates. Bioprocess Biosyst Eng. 2012;32:1229–37. doi:10.1007/s00449-012-0710-8.
Berlemont R, Martiny AC. Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microb. 2013;79:1545–54. doi:10.1128/AEM.03305-12.
Bhat MK. Cellulases and related enzymes in biotechnology. Biotechnol Adv. 2000;18:355–83.
Chauhan JS, Bhat AH, Raghava GPS, Rao A. GlycoPP: a Webserver for prediction of N- and O-glycosites in prokaryotic protein sequences. PLoS ONE. 2012;7:e40155. doi:10.1371/journal.pone.0040155.
Chauvaux S, Beguin P, Aubert JP, Bhat KM, Gow LA, Wood TM, Bairoch A. Calcium-binding affinity and calcium-enhanced activity of Clostridium thermocellum endoglucanase D. Biochem J. 1990;165:261–5.
Cheng Y, Ko T, Huang J, Wu T, Lin C, Luo W, Li Q, Ma Y, Huang C, Wang A, Liu J, Guo R. Enhanced activity of Thermotoga maritima cellulase 12A by mutating a unique surface loop. Appl Microbiol Biotechnol. 2012;95:661–9. doi:10.1007/s00253-011-3791-4.
Chir JL, Wan CF, Chou CH, Wu AT. Hydrolysis of cellulose in synergistic mixtures of β-glucosidase and endo/exocellulase Cel9A from Thermobifida fusca. Biotechnol Lett. 2011;33:777–82. doi:10.1007/s10529-010-0500-9.
Delalibera I, Handelsman JO, Raffa KF. Contrasts in cellulolytic activities of gut microorganisms between the wood borer, Saperda vestita (Coleoptera: Cerambycidae), and the bark beetles, Ips pini and Dendroctonus frontalis (Coleoptera: Curculionidae). Environ Entomol. 2005;34:541–7. doi:10.1603/0046-225X-34.3.541.
Doi RH, Kosugi A. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat Rev Microbiol. 2004;2:541–51. doi:10.1038/nrmicro925.
Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–16. doi:10.1006/jmbi.2000.3903.
Engel P, Moran NA. The gut microbiota of insects-diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. doi:10.1111/1574-6976.12025.
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy Server. In: Walker JM, editor. The proteomics protocols handbook. New York City: Humana Press; 2005. p. 571–607.
Goksoyr J. Cellulases from Sporocytophaga myxococcoides. Methods Enzymol. 1988;160:338–42.
Harshvardhan K, Mishra A, Jha B. Purification and characterization of cellulase from a marine Bacillus sp. H1666: a potential agent for single step saccharification of seaweed biomass. J Mol Catal B. 2013;93:51–6. doi:10.1016/j.molcatb.2013.04.009.
Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–16. doi:10.1042/bj2800309.
Hu X, Yu J, Wang C, Chen H. Cellulolytic bacteria associated with the gut of Dendroctonus armandi larvae (Coleoptera: Curculionidae: Scolytinae). Forest. 2014;5:455–65. doi:10.3390/f5030455.
Irwin D, Shin DH, Zhang S, Barr BK, Sakon J, Karplus PA, Wilson DB. Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J Bacteriol. 1998;180:1709–14.
Juturu V, Wu JC. Microbial cellulases: engineering, production and applications. Renew Sust Energ Rev. 2014;33:188–203. doi:10.1016/j.rser.2014.01.077.
Kim JO, Park SR, Lim WJ, Ryu SK, Kim MK, An CL, Cho SJ, Park YW, Kim JH, Yun HD. Cloning and characterization of thermostable endoglucanase (Cel8Y) from the hyperthermophilic Aquifex aeolicus VF5. Biochem Biophys Res Commun. 2000;279:420–6. doi:10.1006/bbrc.2000.3956.
Kim JY, Hur SH, Hong JH. Purification and characterization of an alkaline cellulase from a newly isolated alkalophilic Bacillus sp. HSH-810. Biotechnol Lett. 2005;27:313–6. doi:10.1007/s10529-005-0685-5.
Kim N, Choo YM, Lee KS, Hong SJ, Seol KY, Je YH, Sohn HD, Jin BR. Molecular cloning and characterization of a glycolyl hydrolase family 9 cellulase distributed throughout the digestive tract of the cricket Teleogryllus emma. Comp Biochem Phys B. 2008;150:368–76. doi:10.1016/j.cbpb.2008.04.005.
Koo HM, Song SH, Pyun YR, Kim YS. Expression and characterization of CMCax having β-1,4-endoglucanase activity from Acetobacter xylinum. J Biochem Mol Biol. 1998;31:53–7.
Kupsky L, Pagnussatt FA, Buffon JG, Furlong EB. Endoglucanase and total cellulase from newly isolated Rhizopus oryzae and Trichoderma reesei: production, characterization, and thermal stability. Appl Biochem Biotech. 2014;172:458–68. doi:10.1007/s12010-013-0518-2.
Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi:10.1038/227680a0.
Lee YJ, Kim BK, Lee BH, Jo KI, Lee NK, Chung CH, Lee YC, Lee JW. Purification and characterization of cellulase produced by Bacillus amyoliquefaciens DL-3 utilizing rice hull. Bioresource Technol. 2008;99:378–86. doi:10.1016/j.biortech.2006.12.013.
Lowry OH, Rosenbrought NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–75.
Lyo AH, Forsberg CW. Endoglucanase from Fibrobacter succinogenes S85 belong to a class of enzymes characterized by a basic C-terminal domain. Can J Microbiol. 1996;42:934–43.
Mazur O, Zimmer J. Apo- and cellopentose-bound structures of the bacterial cellulose synthase subunit BcsZ. J Biol Chem. 2011;286:17601–6. doi:10.1074/jbc.M111.227660.
Medie FM, Davies GJ, Drancourt M, Henrissat B. Genome analyses highlight the different biological roles of cellulases. Nat Rev Microbiol. 2012;10:227–34. doi:10.1038/nrmicro2729.
Mei HZ, Xia DG, Zhao QL, Zhang GZ, Qiu ZY, Qian P, Lu C. Molecular cloning, expression, purification and characterization of a novel cellulase gene (Bh-EGaseI) in the beetle Batocera horsfieldi. Gene. 2016;576:45–51. doi:10.1016/j.gene.2015.09.057.
Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–8.
Morales-Jiménez J, Zúñiga G, Villa-Tanaca L, Hernández-Rodríguez C. Bacterial community and nitrogen fixation in the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Curculionidae: Scolytinae). Microb Ecol. 2009;58:879–91. doi:10.1007/s00248-009-9548-2.
Morales-Jiménez J, Zúñiga G, Ramírez-Saad HC, Hernández-Rodríguez C. Gut-associated bacteria throughout the life cycle of the bark beetle Dendroctonus rhizophagus Thomas and Bright (Curculionidae: Scolytinae) and their cellulolytic activities. Microb Ecol. 2012;64:268–78. doi:10.1007/s00248-011-9999-0.
Ohara H, Noguchi J, Karita S, Kimura T, Sakka K, Ohmiya K. Sequence of egV and properties of EgV, a Ruminococcus albus endoglucanase containing a dockerin domain. Biosci Biotechnol Biochem. 2000;64:80–8. doi:10.1271/bbb.64.80.
Ohmiya Y, Takeda T, Nakamura S, Sakai F, Hayashi T. Purification and properties of a wall-bound endo-1,4-β-glucanase from suspension-cultured poplar cells. Plant Cell Physiol. 1995;36:607–14.
Paradis FW, Zhu H, Krell PJ, Phillips JP, Forsberg W. The xynC gene from Fibrobacter succinogenes S85 codes for a xylanase with two similar catalytic domains. J Bacteriol. 1993;175:7666–72.
Payne CM, Knott BC, Mayes HB, Hansson H, Himmel ME, Sandgren M, Ståhlberg J, Beckham GT. Fungal cellulases. Chem Rev. 2015;115:1308–448. doi:10.1021/cr500351c.
Rajoka MI, Ashraf Y, Khalid AM. Kinetic of improved production and carboxymethyl cellulose hydrolysis by an endo-glucanase from a derepressed mutant of Cellulomonas biazotea. Biotechnol Lett. 2004;26:1329–33.
Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–4.
Romaniec MPM, Fauth U, Kobayashi T, Huskisson NS, Barker PJ, Demain AL. Purification and characterization of a new endoglucanase from Clostridium thermocellum. Biochem J. 1992;283:69–73. doi:10.1042/bj2830069.
Rouvinen J, Bergfors T, Teeri T, Knowles JK, Jones TA. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science. 1990;249:380–6.
Rubini MR, Dillon AJP, Kyaw CM, Faria FP, Poças-Fonseca MJ, Silva-Pereira I. Cloning, characterization and heterologous expression of the first Penicillium echinulatum cellulase gene. J Appl Microbiol. 2010;108:1187–98. doi:10.1111/j.1365-2672.2009.04528.x.
Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, a laboratory manual, vol 1. 2nd ed. New York City: Cold Spring Harbor Laboratory Press; 1989.
Schwarz WH, Bronnenmeier K, Gräbnitz F, Staudenbauer WL. Activity staining of cellulases in polyacrylamide gels containing mixed linkage β-glucans. Anal Biochem. 1987;164:72–7.
Slaytor M. Cellulose digestion in termites and cockroaches: what role do symbionts play. Comp Biochem Phys B. 1992;103:775–84. doi:10.1016/0305-0491(92)90194-V.
Sugimura M, Watanabe H, Lo N, Saito H. Purification, characterization, cDNA cloning and nucleotide sequencing of a cellulase from the yellow-spotted longicorn beetle, Psacothea hilaris. Eur J Biochem. 2003;270:3455–60. doi:10.1046/j.1432-1033.2003.03735.x.
Sun JZ, Scharf ME. Exploring and integrating cellulolytic systems of insects to advance biofuel technology. Insect Sci. 2010;17:163–5. doi:10.1111/j.1744-7917.2010.01348.x.
Taylor TJ, Vaisman II. Discrimination of thermophilic and mesophilic proteins. BMC Struct Biol. 2010;10:S5. doi:10.1186/1472-6807-10-S1-S5.
Vasanthakumar A, Delalibera I, Handelsman J, Klepzig KD, Schloss PD, Raffa KF. Characterization of gut-associated bacteria in larvae and adults of the southern pine beetle, Dendroctonus frontalis Zimmermann. Environ Entomol. 2006;35:1710–7.
Voget S, Steele HL, Streit WR. Characterization of a metagenome-derived halotolerant cellulase. J Biotechnol. 2006;126:26–36. doi:10.1016/j.jbiotec.2006.02.011.
Wang G, Xiaowen Z, Li W, Keke W, Fanglin P, Linsong W. The activity and kinetic properties of cellulases in substrates containing metal ions and acid radicals. Adv Biol Chem. 2012;2:390–5. doi:10.4236/abc.2012.24048.
Watanabe H, Tokuda G. Cellulolytic systems in insects. Annu Rev Entomol. 2010;55:609–32. doi:10.1146/annurev-ento-112408-085319.
Welfle K, Misselwitz R, Welfle H, Politz O, Borriss R. Influence of Ca2+ on conformation and stability of three bacterial hybrid glucanases. Eur J Biochem. 1995;229:726–35. doi:10.1111/j.1432-1033.1995.0726j.x.
Wermelinger B, Seifert M. Analysis of the temperature dependent development of the spruce bark beetle Ips typographus (L.) (Col., Scolytidae). J Appl Ent. 1998;122:185–91. doi:10.1111/j.1439-0418.1998.tb01482.x.
Willis JD, Oppert C, Jurat-Fuentes JL. Methods for discovery and characterization of cellulolytic enzymes from insects. Insect Sci. 2010;17:184–98. doi:10.1016/j.jinsphys.2010.11.019.
Wood SL. The bark and ambrosia beetles of North and Central America (Coleoptera: Scolytidae), a taxonomic monograph. Great Basin Nat Mem. 1982;6:1–1359.
Xu L, Lou Q, Cheng C, Lu M, Sun J. Gut-associated bacteria of Dendroctonus valens and their involvement in verbenone production. Microb Ecol. 2015;70:1012–23. doi:10.1007/s00248-015-0625-4.
Yamane K, Suzuki H, Nisizawa K. Purification and properties of extracellular and cell-bound cellulase components of Pseudomonas fluorescens var cellulosa. J Biochem. 1970;67:19–35.
Yang TC, Legault S, Kayiranga EA, Kumaran J, Ishikawa K, Sung WL. The N-terminal β-sheet of the hyperthermophilic endoglucanase from Pyrococcus horikoshii is critical for thermostability. Appl Environ Microbiol. 2012;78:3059–67. doi:10.1128/AEM.07576-11.
Yennamalli RM, Rader AJ, Wolt JD, Sen TZ. Thermostability in endoglucanases is fold-specific. BMC Struct Biol. 2011;11:1–15. doi:10.1186/1472-6807-11-10.
Zhang G, Li S, Xue Y, Mao L, Ma Y. Effects of salts on activity of halophilic cellulase with glucomannanase activity isolated from alkaliphilic and halophilic Bacillus sp. BG-CS10. Extremophiles. 2012;16:35–43. doi:10.1007/s00792-011-0403-2.
Authors’ contributions
CCR carried out the experiments and wrote the manuscript, ASH and YGH carried out experimental work, FNRO wrote the manuscript, GZ wrote the manuscript, MEHL conceived experiments and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Three anonymous reviewers made valuable suggestions.
Competing interests
The authors declare that they have no competing interests.
Funding information
The project was funded by CINVESTAV-IPN and ICYTDF 45/2012. This work was part of C.C-R postdoctoral residence. C. C-R received a postdoctoral fellowship (202060) from Consejo Nacional de Ciencia y Tecnología, México.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Cano-Ramírez, C., Santiago-Hernández, A., Rivera-Orduña, F.N. et al. Expression, purification and characterization of an endoglucanase from Serratia proteamaculans CDBB-1961, isolated from the gut of Dendroctonus adjunctus (Coleoptera: Scolytinae). AMB Expr 6, 63 (2016). https://doi.org/10.1186/s13568-016-0233-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-016-0233-9