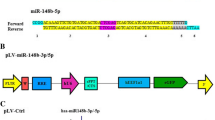
Abstract
Background
The process of bone repair is heavily dependent on the ability of human bone marrow mesenchymal stem cells (hMSCs) to undergo osteogenic differentiation. MicroRNAs have been shown to regulate this osteogenic process. This study aimed to investigate the role of miR-765 in the osteogenic differentiation of hMSCs.
Methods
We transfected hMSCs with lentiviral constructs to knock down or overexpress this miRNA, allowing us to assess its role in osteogenesis via assessing the expression of the relevant markers alkaline phosphatase (ALP), runt-related gene-2 (RUNX-2), and osteocalcin (OCN), with further functional measurements made via quantifying ALP activity and conducting Alizarin Red S staining. The targeting of the 3′-untranslated region (UTR) of BMP6 by miR-765 was examined via luciferase assay. We used hMSCs with altered miR-765 expression to assess p-Smad1/5/9 levels via Western blotting over the course of osteogenic differentiation. We also assessed the osteogenic differentiation of hMSCs in which miR-765 was knocked down and at the same time as a BMP/Smad signaling inhibitor was added to disrupt Smad1/5/9 phosphorylation.
Results
We found miR-765 overexpression to inhibit osteogenesis-associated gene upregulation during osteogenic differentiation of hMSCs, whereas knockdown of this miRNA was associated with increased expression of these genes. Using luciferase reporter assays, we confirmed direct miR-765 binding to the 3′-untranslated region (UTR) of BMP6. We also found that miR-765 overexpression reduced Smad1/5/9 phosphorylation, and knockdown of this miRNA enhanced this phosphorylation on BMP6/Smad1/5/9 signaling. The osteogenic differentiation of hMSCs in which miR-765 had been knocked down was further weakened upon the addition of a BMP/Smad signaling inhibitor relative to miR-765 knockdown alone.
Conclusions
Together, these results thus suggest that miR-765 is able to inhibit hMSC osteogenic differentiation by targeting BMP6 via regulation of the BMP6/Smad1/5/9 signaling pathway. Our findings may offer molecular insights of value for the development of novel therapeutic treatments for bone diseases including osteoporosis.
Similar content being viewed by others
Background
Human mesenchymal stem cells (hMSCs) are self-renewing cells that can be isolated from human bone marrow and which can differentiate into several different cell types [1]. In vivo, these cells can undergo osteoblastic or adipocytic differentiation in response to appropriate stimuli [2,3,4,5]. Disruption of the normal homeostasis between osteoblast and adipocyte differentiation such that relative osteoblast levels are reduced can result in an overall decrease in bone mass as is characteristic of osteoporosis [6]. The specific mechanisms controlling hMSC differentiation have thus been an area of active research, with in vitro expanded hMSCs representing an ideal model system for identifying the regulators of this process [7].
MicroRNAs (miRNAs) are small non-coding RNAs that are able to bind the 3′-untranslated region (UTR) of specific complementary mRNAs, thereby promoting their degradation or translational repression [8]. Several miRNAs have been shown to be linked to osteogenesis, including miR-206, miR-204, and miR-214, all of which can inhibit this process [9,10,11]. In contrast, expression of miR-15b and miR-29b has been found to be pro-osteogenic [12, 13]. This suggests that differential miRNA expression is a key mechanism regulating osteogenic differentiation. The specific regulatory role of miR-765, which is encoded on Chromosome 1 (NC_000001.11; 156936131...156936244) in osteogenesis, however, remains uncertain.
Bone morphogenetic protein (BMP) is a protein that is essential for the normal repair of fractures [14]. Osteoblasts synthesize and secrete these BMP proteins, releasing them in large quantities into the bone matrix environment [15]. In total, there are over 20 BMP family proteins, and they are known to be able to strongly promote osteogenesis via direct stimulation of hMSCs leading to their differentiation [16, 17]. Recent studies have explored the role of miRNAs in the regulation of BMPs within osteoblasts [18,19,20,21]. While BMP6 has been shown to play a key role in the osteogenic process, as has miR-765 which binds to the BMP6 mRNA, the specific underlying signaling mechanisms remain unclear.
In the present study, we assessed the importance of miR-765 in hMSC osteoblastic differentiation, revealing this miRNA to control osteogenesis by targeting BMP6 via regulation of the BMP6/Smad1/5/9 signaling pathway.
Materials
Cell culture and osteogenic differentiation
hMSCs (HUXMA-01001, Cyagen Biosciences, China; cell lot no. of three donors: 150724I31, 160202I31, and 161125R41) were phenotyped via flow cytometry and were confirmed to be ≥ 95% CD73-, CD90-, and CD105-positive, as well as CD11b-, CD19-, CD45-, CD34-, and CD HLA-DR-negative (≤ 5%) (Additional file 1). These cells were initially plated at 5 × 104 cells/cm2 in oriCellTM human Mesenchymal Stem Cell growth media (HUXMA-90011, Cyagen Biosciences) supplemented with 10% FBS, glutamine, and penicillin/streptomycin in a 5% CO2 humidified incubator at 37 °C with 95% air. Every 3–4 days, 0.25% trypsin-EDTA (Gibco; Thermo Fisher Scientific, Inc.) was used to detach cells, which were then passaged. All experiments were conducted with cells from passages 3–6.
To induce osteogenic differentiation, cells were allowed to grow until 70% confluent at which time they were treated with 50 mM ascorbic acid, 10 mM β-glycerophosphate, and 100 nM dexamethasone (all from Sigma-Aldrich, St. Louis, MO) for 20 days, with media being changed every 3 days.
Lentiviral transfection
In order to overexpress or knock down miR-765, appropriate lentiviral vectors were obtained from Shanghai Genechem Co., Ltd. A short hairpin RNA (shRNA) targeting human miR-765 was designed (target sequence, 5′-CATCACCTTCCTTCTCCTCCA-3′). In addition, a negative control (NC) shRNA was used (sequence, 5′-TTCTCCGAACGTGTCACGT-3′). Chemically synthesized DNA oligonucleotides (Shanghai Genechem Co., Ltd.) were respectively cloned into the pGV280 (for shRNA) and pGV369 (for overexpression)-green fluorescent protein (GFP) vector lentiviruses (Shanghai Genechem Co., Ltd.). Cells were transfected to either overexpress miR-765, overexpress a control construct, express a miR-765-specific shRNA, or express a control shRNA. Lentiviral titers were assessed via serial dilution, and hMSCs were added to 6-well plates at 5 × 104 cells/cm2 and allowed to grow until 20–30% confluent. At that time, 1 × 108 TU/ml lentivirus (in 10 μl) was added to cells along with 5 μg/ml polybrene and complete medium. Cells were then allowed to incubate for 10 h at 37 °C, after which fresh medium was added and cells were grown for 72 h. After that time, antibiotic selection was conducted by adding 0.5 μg/ml puromycin to cells, replacing medium every 1–2 days for 6 days in total until surviving cells appeared to proliferate readily.
Alizarin Red S staining
Cells were washed thrice using PBS and were then fixed for 20 min with cold ethanol (95%) and air dried. Next, 0.5% ammonium hydroxide (Sigma-Aldrich, MO) was generated through appropriate dilution of a 30% stock solution in dH2O. Then, Alizarin Red S (ARS) (Sigma-Aldrich, MO) was dissolved in dH2O at a 40-mM concentration, and the ammonium hydroxide solution was used to achieve a pH between 4.1 and 4.3. This ARS solution was then used to stain samples for 30 min at room temperature, followed by three washes using dH2O. Next, 10% (w/v) cetylpyridinium chloride (Sigma-Aldrich, MO) was prepared in 0.1 M sodium phosphate buffer (pH 7.0), and this was used for destaining. Samples were then analyzed based on their absorbance at 560 nm.
Assessment of ALP activity
A commercial ALP Detection Kit (Nanjing Jiancheng Bioengineering Ltd., Nanjing, China) was utilized based on provided protocols. Cells were first frozen and thawed to room temperature four times in order to mediate ALP release. Cell lysates were then added to 96-well plates and combined for 30 min with the ALP substrate at 37 °C, after which a stop buffer was added. Sample absorbance at 560 nm was then assessed via microplate reader (Biorad, Hercules, CA) to measure the enzymatic hydrolysis of the p-nitrophenyl phosphate substrate.
Quantitative real-time PCR
TRIzol (Invitrogen) was used for total RNA extraction based on provided protocols. A Reverse Transcription System and Oligo (dT) was used to generate cDNA based on provided directions (Thermo Scientific). A stem-loop real-time PCR miRNA kit (Ribobio, China) was used for quantifying miR-765 expression, with U6 as an internal control using primers from Ribobio. For mRNA measurements, β-actin served as a normalization control. Primers used herein are shown in Table 1. A SYBR Premix Ex Taq kit (TOYOBO, Japan) was used for conducting qRT-PCR reactions with a 7500 Real-Time PCR System (ABI, Foster, CA, USA), and the 2−ΔΔCT method was used for quantifying gene expression.
Western blotting
Cells were lysed using RIPA buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mmol/l sodium orthovanadate, 1% sodium deoxycholate, 1 mM EDTA, 2 μg/ml leupeptin, 50 mmol/l sodium fluoride). Protein samples were boiled in 5× SDS sample buffer for 5 min, and 15 μg of each samples was separated via 10% SDS-PAGE and transferred onto PVDF membranes (EMD Millipore). Next, 5% skim milk was used to block membranes for 120 min at room temperature, and blots were then probed at 4 °C overnight using the following antibodies: rabbit anti-RUNX2 (ab23981), rabbit anti-OCN (ab133612), rabbit anti-BMP6 (AF5196), rabbit anti-Phospho-Smad1/5/9 (AF8313), rabbit anti-Smad1/5/9 (AF0614), and mouse anti-β-actin (1:2000; ab173838). All rabbit antibodies were diluted 1:1000. Blots were then probed at room temperature for 60 min with an HRP-conjugated secondary antibody specific for either mouse IgG or rabbit IgG (1:5000; Cell Signaling Technology). Protein was then visualized via enhanced chemiluminescence (BeyoECL Plus; Beyotime Institute of Biotechnology).
miRNA target gene prediction
The TargetScan 6.2, PicTar, and miRBase 21 algorithms were used as a means of identifying possible miR-765 target genes.
Luciferase reporter assay
A Dual-Luciferase Reporter Assay System (pGL3 vector; Promega, USA) was used to validate BMP6 as a miR-765 target gene. The BMP6 3′-UTR region containing the putative miRNA binding site of interest was added to the pGL3 vector downstream of luciferase, resulting in the production of either a pGL3-wild-type (WT) or a pGL3-mutant (MUT) vector, with DNA sequencing used to confirm vector construction. These vectors were then transfected into 293T cells either with or without a miR-765 mimic. After 48 h, luciferase activity was assessed based on provided instructions, with Renilla luminescence used to normalize luciferase activity relative to miRNA-transfected controls.
Statistical analysis
Data are means ± standard deviation of experiments repeated three times. SPSS (v16.0; SPSS, Inc.) was used to compare data via one-way ANOVAs, with P < 0.05 as the significance threshold.
Results
miR-765 expression decreases during osteogenic differentiation
We first found that miR-765 expression gradually decreases over the course of osteogenesis, indicating that it may play a key role in regulating hMSC osteogenic differentiation (Fig. 1).
Generation of hMSCs with altered miR-765 expression
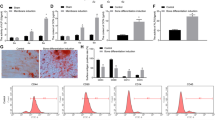
We next generated cells stably expressing vectors that led them to either overexpress miR-765 or an shRNA targeting this miRNA, or to express control vectors. Following puromycin selection, we observed good growth and GFP fluorescence from surviving cells consistent with successful stable transfection (Fig. 2a). We then verified that the lentiviral transfection was successful via qRT-PCR measurement of miR-765, revealing the cells overexpressing miR-765 to have a > 700-fold increase in the expression of this miRNA relative to negative control cells (Fig. 2b).
Stably infected hMSC identification. a Infected cells were assessed via both light and fluorescent microscopy(4×); scale bar, 100 μm. A single representative image is shown. b miR-765 expression was assessed via qRT-PCR. All values are expressed as means ± SD (X ± SD, n = 3). **P < 0.01 vs. negative control
miR-765 inhibits the osteogenic differentiation of hMSCs
To explore how miR-765 influences osteogenic differentiation, we assessed osteogenesis in hMSCs in which miR-765 was knocked down or overexpressed on days 0, 10, and 20 following the addition of osteogenic medium. When we conducted Alizarin Red S staining on days 10 and 20, we found that miR-765 overexpression was associated with reduced staining intensity relative to control cells, suggesting decreased matrix mineralization (Fig. 3a). In contrast, cells in which miR-765 had been knocked down exhibited more intense staining consistent with increased mineralization (Fig. 3b). Differences in mineralization staining intensity were significant among these groups of cells at both days 10 and 20 (Fig. 3c, d), suggesting that miR-765 plays a key role in regulating the osteogenesis of hMSCs.
miR-765 inhibits hMSC osteogenic differentiation. Alizarin Red S staining was employed as a means of assessing matrix mineralization. Scale bar, 50 μm. a miR-765 overexpression was associated with decreased matrix mineralization. b Cell in which miR-765 had been knocked down exhibited more intense staining consistent with increased mineralization. c, d Differences in mineralization staining intensity were significant among these groups of cells. Data are expressed as means ± SD (X ± SD, n = 3). **P < 0.01 vs. negative controls, respectively
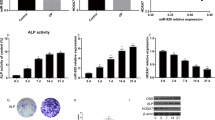
To further explore this process, we next measured expression of the osteogenic markers RUNX2 and OCN via qRT-PCR and Western blotting. We found that miR-765 overexpression was associated with a significant repression of RUNX2 and OCN expression, whereas knockdown of this miRNA was associated with increased expression of both of these genes (Fig. 4a, b). We additionally found miR-765 overexpression to impair ALP activity, whereas this activity was enhanced in cells in which this miRNA had been knocked down (Fig. 4c). This suggests that miR-765 plays a key role in hMSC osteogenesis.
miR-765 inhibits hMSC osteogenic differentiation. a qRT-PCR was used to compare RUNX2, OCN, and ALP expression in the different groups. b RUNX2 and OCN protein levels were assessed via Western blotting. c ALP activity was quantified. Data are expressed as means ± SD (X ± SD, n = 3). **P < 0.01 vs. negative controls, respectively. Note: NC: negative control; OE: miR-765 overexpression; sh: shRNA
miR-765 and BMP6 expression are negatively correlated with one another during osteogenic differentiation
BMP6 as a putative target gene of miR-765, we next assessed the relative expression of these two factors in the context of osteogenesis via qRT-PCR. We found that there was a clear negative correlation between the expression of miR-765 and BMP6 during hMSC osteogenic differentiation (Fig. 5).
BMP6 is a direct target of miR-765
We next sought to directly assess the ability of miR-765 to regulate hMSC osteogenic differentiation via targeting BMP6. We therefore assessed the impact of miR-765 on BMP6 expression at different stages of the osteogenic differentiation process (Fig. 6a). Based on TargetScan analyses, we identified a miR-765 targeting site located within the BMP6 3′-UTR (Fig. 6b). To confirm that this was indeed a binding site for this miRNA, we utilized a standard luciferase reporter assay. This assay confirmed that there was a 50% reduction in luciferase activity in cells expressing the WT version of this sequence that had been transfected with miR-765 relative to those that had not. In contrast, cells expressing the mutated version of this construct did not exhibit any reduction in luciferase activity upon miR-765 transfection (Fig. 6c).
miR-765 targets the 3′-UTR of BMP6. a The ability of miR-765 to impact BMP6 expression over the course of osteogenic differentiation in hMSCs following stable lentiviral transfected was assessed. b The miR-765 binding region in the BMP6 3′-UTR. c Luciferase activity assay. All values are expressed as means ± SD (X ± SD, n = 3). **P < 0.01 vs. negative controls, respectively. Note: NC: negative control; OE: miR-765 overexpression; sh: shRNA
miR-765 derepresses BMP6/Smad1/5/9 signaling during osteogenic differentiation
To assess whether miR-765 expression has any influence on Smad signaling in the context of osteogenesis, we used our stable hMSC constructs to assess p-Smad1/5/9 levels by Western blotting over the course of osteogenic differentiation. We found that miR-765 overexpression was associated with reduced p-Smad1/5/9 levels, whereas miR-765 knockdown was associated with increased p-Smad1/5/9 levels relative to appropriate controls (Fig. 9a, b).
Cells in which miR-765 had been knocked down were treated with 100 nM LDN-193189 (Selleck, USA), a small molecule inhibitor of BMP type I receptor that was used to specifically inhibit BMP/Smad signaling, in order to reduce p-Smad1/5/9 levels in these cells (Fig. 9c), revealing that hMSC osteogenic differentiation in cells in which miR-765 had been knocked down was weaker as compared with that in cells in which miR-765 had been knocked down in the absence of inhibitor addition (Figs. 7, 8). These results thus further revealed that BMP6/Smad1/5/9 is downstream of miR-765.
The osteogenic differentiation of hMSCs in which miR-765 was knocked down and at the same time as a BMP/Smad signaling inhibitor was added to disrupt Smad1/5/9 phosphorylation. Alizarin Red S staining was employed as a means of assessing matrix mineralization. Scale bar, 50 μm. a By adding a BMP/Smad signaling inhibitor, the osteogenic differentiation of hMSCs in which miR-765 was also knocked down was associated with decreased matrix mineralization relative to cells in which miR-765 was knocked down but no inhibitor was added. b Differences in mineralization staining intensity were significant between shRNA+inhibitor and shRNA groups. Data are expressed as means ± SD (X ± SD, n = 3). **P < 0.01 vs. shRNA+inhibitor, respectively
The osteogenic differentiation of hMSCs in which miR-765 was knocked down and at the same time as a BMP/Smad signaling inhibitor was added to block Smad1/5/9 phosphorylation. a qRT-PCR was used to compare RUNX2, OCN, and ALP expression in the different groups. b RUNX2 and OCN protein levels were assessed via Western blotting. c ALP activity was quantified. Data are expressed as means ± SD (X ± SD, n = 3). **P < 0.01 vs. shRNA+inhibitor, respectively. Note: sh: shRNA; sh+inh: sh+inhibitor
Discussion
Recent work has clearly highlighted the role of endogenous miRNAs as key regulators of stem cell differentiation and renewal [22, 23]. Consistent with this, in the present study, we observed the gradual downregulation of miR-765 in hMSCs as they underwent osteogenic differentiation (Fig. 1). This suggested that this miRNA may play a key role in this osteogenic process, leading us to generate hMSCs stably overexpressing miR-765 or an shRNA targeting this miRNA (Fig. 2a, b). When we used these cells to assess changes in osteogenesis, we found miR-765 overexpression to significantly impair ALP activity and matrix mineralization, whereas knockdown of this miRNA had the opposite effect (Fig. 3 and Fig. 4c). Consistent with this, miR-765 overexpression impaired the expression of osteogenic markers in hMSCs during osteogenic differentiation, while knocking down this miRNA increased the expression of these markers in this same context (Fig. 4a, b). This thus indicated that miR-765 acts to negatively regulate the osteogenic differentiation of hMSCs.
Previous work has shown that miR-765 plays a role in many contexts, including arteriosclerosis, multiple myeloma, tongue squamous carcinoma, hepatoma cell proliferation, osteosarcoma cell migration, and neural stem cell differentiation [24,25,26,27,28,29]. The role of miR-765 in hMSC osteogenic differentiation, however, has not been clearly confirmed in previous studies. Our study was able to confirm a central role for miR-765 in this process.
Developing a better understanding of how miR-765 regulates osteogenesis has the potential to highlight novel therapeutic strategies for treating osteoporosis. We therefore investigated potential miR-765 target genes, identifying and confirming BMP6 as a miR-765 target gene, the expression of which was negatively correlated with that of miR-765 (Fig. 5).
To confirm that BMP6 was truly a functional target of miR-765, we next assessed changes in BMP6 expression upon modulation of miR-765 expression. We found that miR-765 overexpression was linked to a significant drop in BMP6 expression, whereas knockdown of this miRNA markedly enhanced BMP6 expression (Fig. 6a). miRNAs inhibit target mRNA translation via binding to complementary sequences in the 3′-UTR region of these mRNAs, thereafter mediating their degradation [30]. We therefore next used a standard luciferase reporter assay to validate the presence of a direct interaction between miR-765 and the BMP6 3′-UTR, with our results confirming BMP6 to be a miR-765 target (Fig. 6b, c). This is the first study we are aware of to have demonstrated that miR-765 inhibits the osteogenic differentiation of hMSC via targeting BMP6.
BMP6 is a protein that plays key roles in the formation of bone and in the regulation of osteoporosis, bone cancer, and fracture healing [31,32,33,34]. BMP6 is a member of the transforming growth factor β (TGF-β) superfamily, which is able to bind the BMPR class II receptor and then recruits a BMPRI-type receptor ultimately leading to Smad1/5/9 protein phosphorylation and activation. Upon activation, p-Smad1/5/9 is then able to cooperate with Smad4 to translocate into the nucleus wherein it can regulate the expression of genes related to diverse processes such as differentiation, growth, proliferation, migration, and apoptosis [16, 35,36,37]. This thus suggested that this BMP6/Smad1/5/9 pathway is likely to be a key regulatory mechanism in the context of hMSC osteogenic differentiation. As this pathway has not been studied in this context previously, we chose to examine it in depth, revealing that miR-765 overexpression was associated with reduced Smad1/5/9 phosphorylation upon osteogenic differentiation, whereas miR-765 knockdown significantly enhanced p-Smad1/5/9 levels, suggesting that this BMP6/Smad1/5/9 signaling pathway plays a key role in osteogenic differentiation and that miR-765 controls its activation (Fig. 9a, b). Furthermore, by adding a BMP/Smad signaling inhibitor to reduce p-Smad1/5/9 levels (Fig. 9c), we were able to demonstrate that the osteogenic differentiation of hMSCs in which miR-765 was also knocked down was weaker than in cells in which miR-765 was knocked down but no inhibitor was added (Figs. 7 and 8). These results thus further confirmed that BMP6/Smad1/5/9 is downstream of miR-765, with miR-765 controlling the activation of this pathway.
miR-765 suppresses BMP6/Smad1/5/9 signaling to inhibit osteogenesis. Western blotting was used to measure p-Smad1/5/9 levels, with β-actin as a loading control. a miR-765 overexpression was associated with reduced Smad1/5/9 phosphorylation upon osteogenic differentiation. b miR-765 knockdown significantly enhanced p-Smad1/5/9 levels. c By adding a BMP/Smad signaling inhibitor to reduced p-Smad1/5/9 levels. BMP6/Smad1/5/9 was shown to be downstream of miR-765, with miR-765 controlling the activation of this pathway. Data are expressed as means ± SD (X ± SD, n = 3). **P < 0.01 vs. negative controls or shRNA+inhibitor, respectively. Note: NC: negative control; OE: miR-765 overexpression; sh: shRNA; sh+inh: sh+inhibitor
Conclusions
In summary, our results indicate that miR-765 is able to suppress hMSC osteogenic differentiation through targeting BMP6, thereby regulating the BMP6/Smad1/5/9 signaling pathway. These findings may offer novel opportunities to develop therapeutic regimens for the treatment of osteoporosis or other bone diseases.
Availability of data and materials
All data generated and/or analyzed during this study are included in this published article.
Abbreviations
- ALP:
-
Alkaline phosphatase
- BMP6:
-
Bone morphogenetic protein 6
- hMSCs:
-
Human bone marrow-derived mesenchymal stem cells
- miRNA:
-
MicroRNA
- OCN:
-
Osteocalcin
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- RUNX2:
-
Runt-related gene-2
- UTR:
-
Untranslated region
References
Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–7.
Barry F, Boynton RE, Liu B, Murphy JM. Chondrogenic differentiation of mesenchymal stem cells from bone marrow: differentiation-dependent gene expression of matrix components. Exp Cell Res. 2001;268(2):189–200.
Arinzeh TL. Mesenchymal stem cells for bone repair: preclinical studies andpotential orthopedic applications. Foot Ankle Clin. 2005;10(4):651–65.
Helder MN, Knippenberg M, Klein-Nulend J, Wuisman PI. Stem cells from adipose tissue allow challenging new concepts for regenerative medicine. Tissue Eng. 2007;13(8):1799–808.
Rosen C, Karsenty G, MacDougald O. Foreword: interactions between bone and adipose tissue and metabolism. Bone. 2012;50(2):429.
Scheideler M, Elabd C, Zaragosi LE, Chiellini C, Hackl H, Sanchez-Cabo F, Yadav S, Duszka K, Friedl G, Papak C. Comparative transcriptomics of human multipotent stem cells during adipogenesis and osteoblastogenesis. BMC Genomics. 2008;9:340.
Okolicsanyi RK, Camilleri ET, Oikari LE, Yu C, Cool SM, van Wijnen AJ, Griffiths LR, Haupt LM. Human mesenchymal stem cells retain multilineage differentiation capacity including neural marker expression after extended in vitro expansion. PLoS One. 2015;10(9):e0137255.
Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5.
Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28(2):357–64.
Sato MM, Nashimoto M, Katagiri T, Yawaka Y, Tamura M. Bone morphogenetic protein-2 downregulates miR-206 expression by blocking its maturation process. Biochem Biophys Res Commun. 2009;383(1):125–9.
Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A, Li D, Hou Z, Lv K, Kan G, et al. miR-214 targets ATF4 to inhibit bone formation. Nat Med. 2013;19(1):93–100.
Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284(25):15676–84.
Vimalraj S, Partridge NC, Selvamurugan N. A positive role of microRNA-15b on regulation of osteoblast differentiation. J Cell Physiol. 2014;229(9):1236–44.
Dumic-Cule I, Peric M, Kucko L, Grgurevic L, Pecina M, Vukicevic S. Bone morphogenetic proteins in fracture repair. Int Orthop. 2018;42(11):2619–26.
Jain AP, Pundir S, Sharma A. Bone morphogenetic proteins: the anomalous molecules. J Indian Soc Periodontol. 2013;17(5):583–6.
Beederman M, Lamplot JD, Nan G, Wang J, Liu X, Yin L, Li R, Shui W, Zhang H, Kim SH, et al. BMP signaling in mesenchymal stem cell differentiation and bone formation. J Biomed Sci Eng. 2013;6(8A):32–52.
Jung T, Lee JH, Park S, Kim YJ, Seo J, Shim HE, Kim KS, Jang HS, Chung HM, Oh SG, et al. Effect of BMP-2 delivery mode on osteogenic differentiation of stem cells. Stem Cells Int. 2017;2017:7859184.
Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, Lian JB, Stein GS, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc NatlAcad Sci USA. 2008;105(37):13906–11.
Kureel J, Dixit M, Tyagi AM, Mansoori MN, Srivastava K, Raghuvanshi A, Maurya R, Trivedi R, Goel A, Singh D. miR-542-3p suppresses osteoblast cell proliferation and differentiation, targets BMP-7 signaling and inhibits bone formation. Cell Death Dis. 2014;5:e1050.
Zhang W, Zhang L, Zhou Y, Ji X, Liu J, Liu D, Yin P, Peng Y, Hao M, Zhang L, et al. Synergistic effects of BMP9 and miR-548d-5p on promoting osteogenic differentiation of mesenchymalstem cells. Biomed Res Int. 2015;2015:309747.
Chen C, Tang Z, Song Q, Yang M, Shi Q, Weng Y. Downregulated microRNA-23b promotes BMP9-mediatedosteogenesis in C2C12 myoblast cells by targeting Runx2. Mol Med Rep. 2016;13(3):2492–8.
Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10(2):116–25.
Lakshmipathy U, Hart RP. Concise review: MicroRNA expression in multipotent mesenchymal stromal cells. Stem Cells. 2008;26(2):356–63.
Liao YC, Wang YS, Hsi E, Chang MH, You YZ, Juo SH. MicroRNA-765 influences arterial stiffness through modulating apelin expression. Mol Cell Endocrino. 2015;411:11–9.
Li S, Zhao W, Xu Q, Yu Y, Yin C. MicroRNA-765 regulates neural stem cell proliferation and differentiation by modulating Hes1 expression. Am J Transl Res. 2016;8(7):3115–23.
Xie BH, He X, Hua RX, Zhang B, Tan GS, Xiong SQ, Liu LS, Chen W, Yang JY, Wang XN, et al. Mir-765 promotes cell proliferation by downregulating INPP4B expression in human hepatocellular carcinoma. Cancer Biomark. 2016;16(3):405–13.
Ding J, Yang C, Yang S. LINC00511 interacts with miR-765 and modulates tongue squamous cell carcinoma progression by targeting LAMC2. J Oral Pathol Med. 2018;47(5):468–76.
Yan L, Wu X, Liu Y, Xian W. LncRNA Linc00511 promotes osteosarcoma cell proliferation and migration through sponging miR-765. J Cell Biochem. 2018. https://doi.org/10.1002/jcb.27999.
Long S, Long S, He H, Chen G. MicroRNA-765 is pregulated in multiple myeloma and serves an oncogenic role by directly targeting SOX6. Exp Ther Med. 2019;17(6):4741–7.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33.
Dai J, Keller J, Zhang J, Lu Y, Yao Z, Keller ET. Bone morphogenetic protein-6 promotes osteoblastic prostate cancer bone metastases through a dual mechanism. Cancer Res. 2005;65(18):8274–85.
Simic P, Buljan-Culej J, Orlic I, Grgurevic L, Draca N, Spaventi R, Vukicevic S. Systemically administered bone morphogenetic protein-6 restores bone in aged ovariectomized rats by increasing bone formation and suppressing bone resorption. J Biol Chem. 2006;281(35):25509–21.
Kayabaşi GK, Aydin RS, Gümüşderelioğlu M. In vitro chondrogenesis by BMP6 gene therapy. J Biomed Mater Res A. 2013;101(5):1353–61.
Zhang Y, Zhang DY, Zhao YF, Wang J, He JW, Luo J. Analysis and characterization of the functional TGF-β receptors required for BMP 6-induced osteogenic differentiation of mesenchymal progenitor cells. BMB Rep. 2013;46(2):107–12.
Ebisawa T, Tada K, Kitajima I, Tojo K, Sampath TK, Kawabata M, Miyazono K, Imamura T. Characterization of bone morphogenetic protein-6 signaling pathways in osteoblast differentiation. J Cell Sci. 1999;112(Pt 20):3519–27.
Hakki SS, Bozkurt B, Hakki EE, Kayis SA, Turac G, Yilmaz I, Karaoz E. Bone morphogenetic protein-2, -6, and -7 differently regulate osteogenic differentiation of human periodontal ligament stem cells. J Biomed Mater Res B Appl Biomater. 2014;102(1):119–30.
Li CJ, Madhu V, Balian G, Dighe AS, Cui Q. Cross-talk between VEGF and BMP-6 pathways accelerates osteogenic differentiation of human adipose-derived stem cells. J Cell Physiol. 2015;230(11):2671–82.
Acknowledgements
Not applicable.
Individual person’s data
Not applicable.
Funding
This study was supported by the National Nature Science Foundation of China (no. 81860260).
Author information
Authors and Affiliations
Contributions
TW and CZ conceived this study. CW, JL, HY, XZ, JZ, XW, SH, XC, and WL performed the experiments, collected data, and performed the data analysis. XX and BM helped perform the experiments. TW wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Dentification of hMSCs via flow cytometry.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, T., Zhang, C., Wu, C. et al. miR-765 inhibits the osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting BMP6 via regulating the BMP6/Smad1/5/9 signaling pathway. Stem Cell Res Ther 11, 62 (2020). https://doi.org/10.1186/s13287-020-1579-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13287-020-1579-0