Abstract
Background and purposes
Brain morphometry is an important assessment technique to assess certain morphological brain features of various brain regions, which can be quantified in vivo by using high-resolution structural magnetic resonance (MR) imaging. This study aims to investigate the effect of different types of pulse sequence on regional corpus callosum (CC) morphometry analysis.
Materials and methods
Twenty-one healthy volunteers were scanned twice on the same 3T MRI scanner (Magnetom Trio, Siemens, Erlangen, Germany) equipped with an 8-channel head coil. Two different MR pulse sequences were applied to acquire high-resolution 3D T1-weighted images: magnetization-prepared rapid gradient-echo (MP-RAGE) and modified driven equilibrium Fourier transform (MDEFT) pulse sequence. Image quality measurements such as SNR, contrast-to-noise ratio, and relative contrast were calculated for each pulse sequence images independently. The values of corpus callosum volume were calculated based on the vertex of reconstructed surfaces. The paired dependent t test was applied to compare the means of two matched groups.
Results
Three sub-regional CC, namely anterior, mid-anterior, and posterior, resulted in an estimated volume difference between MDEFT and MP-RAGE pulse sequences. Central and mid-posterior sub-regional CC volume resulted in not significant difference between the two named pulse sequences.
Conclusion
The findings of this study demonstrate that combining data from different pulse sequences in a multisite study could make some variations in the results.
Similar content being viewed by others
Key points
-
Brain morphometry is an important assessment technique to investigate certain morphological brain features of various brain regions, which can be quantified in vivo by using high-resolution structural magnetic resonance (MR) imaging.
-
Different MR field strengths and pulse sequences could possibly cause some variations on brain morphometric measurements.
-
Combining data from different pulse sequences in a multisite study could make some variations of the results.
-
Strict MR parameter options should be determined carefully by automated brain segmentation software packages in order to avoid any misclassification of voxels that are allocated between surrounding tissue types such as gray matter, white matter, and cerebrospinal fluid.
Introduction
Brain morphometry is an important assessment of certain morphological brain features such as volume, surface, thickness, and shape of various brain regions such as frontal lobe, corpus callosum, and hippocampus that can be measured in vivo by using high-resolution structural magnetic resonance MR imaging technology. It has been used widely to investigate the effect and causes of certain neurological, neurogenerative, psychological, and psychiatric disorders such as epilepsy [1], cognitive impairments [2], autism [3], and schizophrenia [4]. Morphological changes in brain structures could be related to ageing degeneration [5], some neurological disorders [6], and treatment effects [7].
Corpus callosum (CC) is the primary commissural region of the brain consisting of white matter tracts, which connects the right and left hemispheres of the brain, and allows to integrate and transfer the information between the two halves in order to process essential signals such as sensory, motor, and high-level cognitive. CC atrophy is a possible indicator of region- and cell type–specific neuronal degeneration in Alzheimer’s diseases [8]. The relationship between regional microstructural abnormalities of the CC and physical and cognitive disability was studied in the replacing-remitting multiple sclerosis (RRMS) patients, which concludes that regional CC imaging properties differentially explained disability within RRMS patients revealing strong, distinct patterns of correlation with clinical and cognitive status of patients affected by this specific clinical phenotype [9]. Furthermore, regional CC morphometry could demonstrate that specific CC regions may contribute to the cognitive and dysfunction of multiple sclerosis patients [10].
Different MR field strengths, pulse sequences, and parameters could possibly cause some variations on CC morphometric measurements [11,12,13]. MR field strengths for clinical purposes can vary between 0.2 and 3 T, whereas for experimental purposes up to 11 T. The effect of field strength on MR image quality has been investigated widely [11, 13,14,15]. High-field-strength MR imaging (1.5 T and above) is considered to acquire high-quality MR images; however, it does not confer higher accuracy in the diagnosis of multiple sclerosis [16]. Furthermore, a simple change of MR parameters, notably spatial resolution, contrast, and filtering, were found to systematically bias the results of automated brain MRI morphometry [12]. Spatial resolution and modification in contrast resulted in relative estimated volume difference of up to 4.28% in cortical GM and 4.16% in the hippocampus between the same MR pulse sequence type with different parameters (1.0 versus 1.2 mm iso-voxel) [12].
Brain structural morphometry studies require a large sample size, and in order to reduce this effect, it is recommended to optimize the imaging acquisition and analysis protocols [17]. Several methods have been applied to obtain brain morphometry such as manual and automated segmentation [18, 19]. Brain morphometry reproducibility was investigated in multi-center 3T MRI sites, which found that longitudinal analysis yields a consistently improved reproducibility across the various sites relative to the cross-sectional segmentation, reducing the variability by about half in most volumetric estimates and in the entorhinal cortical thickness, while not significantly changing the variability in the rest of cortical structures studied [17].
The aim of this study is to investigate the effect of different types of pulse sequence on regional CC morphometry analysis. This will be justified by comparing the image quality and regional CC volumetric analysis among scans of the same participants who have been scanned at the same field strength of 3 T with different MR pulse sequences (magnetization-prepared rapid gradient-echo (MP-RAGE)/modified driven equilibrium Fourier transform (MDEFT)). The main objective of this study was to identify whether scanning at the same MR field strength with different pulse sequences could affect the reliability of brain volumetric analysis.
Material and methods
Subjects
Twenty-one subjects (15 males and 6 females) with a mean age of 45 years and a standard deviation (SD) of 12 years participated in this study. All participants received the participants’ information sheet and submitted consent forms prior to the examination. This study has been approved by the National Research Ethics Services (NRES) Committee North West-Liverpool Central.
MRI data acquisition
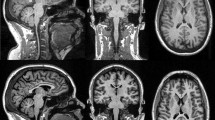
All data were acquired on the same 3T MRI scanner (Magnetom Trio, Siemens, Erlangen, Germany) equipped with an 8-channel head coil. Two different MR pulse sequences were applied to acquire high-resolution 3D T1-weighted images: MP-RAGE and MDEFT pulse sequence (Fig. 1). Scanning parameters for each MR pulse sequence are listed in Table 1. As the acquisition time is quite similar between the two pulse sequences, it is important to mention that no additional acceleration techniques have been applied in this study. MR images were visually inspected in order to approve appropriate image quality and to exclude subjects with visible brain abnormalities.
MRI data analysis
MR image quality
Image quality measurements such as SNR and contrast-to-noise ratio (CNR) were calculated for each pulse sequence images. 3D MP-RAGE and 3D MDEFT images for each participant were acquired during the same session in each scanner, maintaining their alignment. The anatomic region of interests (ROIs) include two GM structures (cortex and hippocampus) and two WM structures (corpus callosum and internal capsule), plus a ROI in artifact-free background to sample noise. The ROIs were drawn using the software ImageJ (National Institutes of Health, Bethesda, MD, USA). SNR values for GM and WM were calculated for each subject by averaging across the pair of ROIs corresponding to the respective tissue type. The CNR was then calculated for each subject by averaging across subjects for each imaging method.
CC Volumetric analysis
Surface-based morphometry (SBM) was analyzed using FreeSurfer software version 5.3. It is an automated procedure that derives morphometric measures from geometric models of the cortical surface. 3D T1-weighted images (MP-RAGE and MDEFT) were corrected for intensity bias. Gray and white matter surfaces were reconstructed by segmenting corrected T1-weighted images into GM and WM surfaces. Intracranial volume (ICV) was calculated by adding the values of gray matter, white matter, and cerebral fluid volumes together. The values of corpus callosum volume were calculated based on the vertex of reconstructed surfaces. These data were then smoothed using 10-mm FWHM Gaussian kernel in surface-space in order to improve SNR and inter-subject registration. Based on the anatomical and histological features, CC were segmented into five main sub-regions: anterior, mid-anterior, central, mid-posterior, and posterior regions (Fig. 2).
Statistical analysis
Normality of residuals was tested using D’Agostino-Pearson normality test in order to quantify how far the distribution is from the Gaussian in terms of asymmetry and shape. Normality test reveals that all data were sampled from a population and follows a Gaussian distribution.
Paired dependent t test was applied to compare the means of two matched groups assuming that the distribution of the SNR and CNR data differences follows a Gaussian distribution. Intraclass correlation coefficient (ICC) is applied to perform inter-rater reliability among the MR pulse sequences. The paired dependent t test was applied as well as compare the CC volume means of the two matched groups assuming that the distribution of the corpus callosum and intracranial volume data differences follows a Gaussian distribution. All data were analyzed using GraphPad Prism for MacOS version 8.
Results
MR image quality
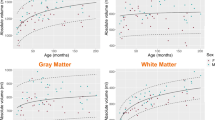
No significant differences were found between the SNR values of the gray matter (GM) regions in the MDEFT and MP-RAGE pulse sequences (t = 0.15, p = 0.87). The mean SNR of the white matter (WM) regions resulted in a significant difference of up to 6% (t = 2.52, p = 0.02) in the MDEFT pulse sequence (121.77 ± 78.45) compared with MP-RAGE pulse sequence (114.47 ± 73.45). Also, the average CNR and relative contrast resulted in significant difference up to 40% and 30% respectively (t = 5.43 and t = 8.08 respectively, p < 0.0001) in MDEFT pulse sequence (57.94 ± 45.38 and 140.8 ± 15.09 respectively) compared with MP-RAGE pulse sequence (34.69 ± 21.85 and 101.1 ± 20.2 respectively) (Table 2, Fig. 3).
Total intracranial volume (ICV) was found significantly higher (p < 0.0001) in the MDEFT pulse sequence (1792.4 ± 197.8 mL) compared with the MP-RAGE pulse sequence (1195.2 ± 133.1 mL) (Fig. 4). Three sub-regional CC volumes, namely anterior (p < 0.0001), mid-anterior (p = 0.003), and posterior (p < 0.0001) were found significantly larger in MDEFT pulse sequence compared with the MP-RAGE pulse sequence. Central and mid-posterior sub-regional CC volume was found no significant difference between the two pulse sequences (Table 3 and Fig. 5).
Reproducibility coefficients for volumetric measurements of CC regions in the two pulse sequences (MDEFT/MP-RAGE) are presented in Table 4. Reliability was very good which ranges between 0.89 and 0.95 for all CC regions in the comparison between MDEFT and MP-RAGE pulse sequences. This reliability test resulted in a statistically significant difference (p < 0.0001) for all the CC regions between the two examined pulse sequences.
Discussion
This study examined the reliability of regional CC morphometry analysis between two different pulse sequence (MDEFT and MP-RAGE) at the same MR field strength (3 T) using an automated procedure derives morphometric measures from geometric models of the cortical surface. There is a significant difference in volumes up to 33% between MDEFT and MP-RAGE when evaluating total ICV that might be resulted in the significant differences of CNR and relative contrast up to 40% and 30% respectively. The volumes of three sub-regions of CC (anterior, mid-anterior, and posterior) were found significant differences across the two examined MR pulse sequences. Reliability measurements were found significantly high in all CC regions across the two examined MR pulse sequences.
In this study, we reported that MDEFT pulse sequence acquires a better SNR in WM, CNR, and relative contrast compared with MP-RAGE pulse sequence, while the SNR in GM seems to be similar between these two pulse sequences, which is consistent to similar study by Tardif et al. who reported that MDEFT pulse sequence achieved the highest CNR between WM and GM, and the lowest GM density variability compared with MP-RAGE and FLASH pulse sequences [20]. A more recent introduced sequence MDEFT was suggested to be more favorable than MP-RAGE at high field strengths [21]. It is characterized by a distinct regional bias in GM density due to the effect of transmission field inhomogeneity on image uniformity combined with spatially variant GM T(1) values and the sequence’s T(1) contrast function [20]. Compared with MP-RAGE, MDEFT was proposed to be relatively insensitive to inhomogeneities in the B1 field [22]. SNR, CNR, and relative contrast values obtained from different pulse sequences (MDEFT/MP-RAGE) at the same field strength of 3 T were different. Relying on the findings of the current study and combining and comparing SNR, CNR, and relative contrast data of different pulse sequences (MDEFT/MP-RAGE) at the same field strength of 3 T appears to be possible. Therefore, the results of this study should be carefully assessed when comparing them to results from other field strengths.
Several methods for examining the brain morphometry are relied on high-resolution T1-weighted using (MP-RAGE) or (MDEFT) pulse sequences [11, 23,24,25]. MP-RAGE offers good contrast between brain tissues: the gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) [26, 27]. It captures high tissue contrast and provides high spatial resolution with whole-brain coverage [28]. On the other hand, 3D MDEFT sequence has been optimized recently for anatomical brain imaging at 1.5 T and 3.0 T [29]. It is proposed that 3D MDEFT lowers its sensitivity to RF inhomogeneity that is particularly essential at high field strengths where RF focusing effects aggravate B1 inhomogeneity, causing major signal non-uniformity in the MR images [30]. It is worth to mention that bias field correction algorithm is an image preprocessing step usually used for segmentation technique in order to correct low frequency intensity non-uniformity present in MR image data and minimizing segmentation error [31, 32]. It was suggested that the MP-RAGE sequence noticeably improved the outlining of GM and WM and significantly reduced flow artifacts [33]. On the other hand, the MDEFT consists of a saturation pulse and an inversion pulse, which is usually used to reduce radio-frequency inhomogeneity sensitivity and improves brain tissue contrast [30]. MDEFT was suggested to be more satisfactory than MP-RAGE at high field strengths [21].
In this study, the same software pipeline (FreeSurfer) was used to quantify the CC volume in order to eliminate the effect of different software pipelines on brain morphometry measurements. In automated FreeSurfer segmentation, the CC was found appropriate for analysis without manual correction [19]. Furthermore, a constant bandwidth was applied in this study as it represents a potential difference from typical clinical conditions, under which the minimum bandwidth for each unit is generally used [13]. Also, a pilot study had been undertaken in order to assess the segmentation quality of each pulse sequence before starting participant recruitment.
As only healthy subjects were recruited in this study, it is difficult to draw a definite conclusion regarding the effect of different pulse sequences on the diagnostic accuracy for some neurological cases. In addition, automatic segmentation performed for CC morphometry analysis may cause some misclassification of some voxels. In other words, given voxels were allocated in GM and classified as WM or CSF because of the limited resolution of MRI scanner. This limitation was solved by visual inspection and manual correction to ensure that voxels are representing the correct brain tissues. The result of this paper may only apply for the same version of the FreeSurfer pipeline, which needs to be compared with the other versions of the FreeSurfer package in order to evaluate the difference of measured volumes between different FreeSurfer package versions.
Conclusion
The findings confirm that the MDEFT pulse sequence acquires a better SNR in the WM, CNR and relative contrast compared with the MP-RAGE pulse sequence, while the SNR in the GM seems to be similar between these two pulse sequences. Relying on the findings of the current study, the variation of brain volumetric measurements between different pulse sequences (MDEFT/MP-RAGE) at the same field strength of 3 T seems to be possible. Combining data from different pulse sequences in a multisite study could make some variations of the results. Therefore, the findings of this study should be carefully assessed when comparing them to results from other field strengths and MRI scanner manufactures. Furthermore, strict MR parameter options should be determined carefully by automated brain segmentation software packages in order to avoid any misclassification of voxels that are allocated between surrounding tissue types such as GM, WM, and CSF. Future studies are required to investigate the effect of other factors such as scanner manufacturers, coil channel selections, and other pulse sequences parameters options on the MR image quality and brain morphometry analysis.
Abbreviations
- CC:
-
Corpus callosum
- CSF:
-
Cerebrospinal fluid
- GM:
-
Gray matter
- ICV:
-
Intracranial volume
- MDEFT:
-
Modified driven equilibrium Fourier transform
- MP-RAGE:
-
Magnetization-prepared rapid gradient-echo
- MR:
-
Magnetic resonance
- NRES:
-
National Research Ethics Services
- RRMS:
-
Replacing-remitting multiple sclerosis
- SBM:
-
Surface-based morphometry
- T:
-
Tesla
- TE:
-
Echo time
- TI:
-
Inversion time
- TR:
-
Repetition time
- WM:
-
White matter
References
Alhusaini S, Kowalczyk MA, Yasuda CL et al (2019) Normal cerebral cortical thickness in first-degree relatives of temporal lobe epilepsy patients. Neurology 92:e351–e358
Kim HJ, Ye BS, Yoon CW et al (2014) Cortical thickness and hippocampal shape in pure vascular mild cognitive impairment and dementia of subcortical type. Eur J Neurol 21:744–751
Schumann CM, Barnes CC, Lord C, Courchesne E (2009) Amygdala enlargement in toddlers with autism related to severity of social and communication impairments. Biol Psychiatry 66:942–949
Welch KA, Stanfield AC, Moorhead TW et al (2010) Amygdala volume in a population with special educational needs at high risk of schizophrenia. Psychol Med 40:945–954
Castellanos FX, Lee PP, Sharp W et al (2002) Developmental trajectories of brain volume abnormalities in children and adolescents with attention-deficit/hyperactivity disorder. JAMA 288:1740–1748
Fotenos AF, Snyder AZ, Girton LE, Morris JC, Buckner RL (2005) Normative estimates of cross-sectional and longitudinal brain volume decline in aging and AD. Neurology 64:1032–1039
Yang C-Y, Liu H-M, Chen S-K, Chen Y-F, Lee C-W, Yeh L-R (2016) Reproducibility of brain morphometry from short-term repeat clinical MRI examinations: a retrospective study. PLoS One 11:–e0146913
Hampel H, Teipel SJ, Alexander GE et al (1998) Corpus callosum atrophy is a possible indicator of region- and cell type-specific neuronal degeneration in Alzheimer disease: a magnetic resonance imaging analysis. Arch Neurol 55:193–198
Caligiuri ME, Barone S, Cherubini A et al (2015) The relationship between regional microstructural abnormalities of the corpus callosum and physical and cognitive disability in relapsing–remitting multiple sclerosis. Neuroimage Clin 7:28–33
Yaldizli Ö, Penner I-K, Frontzek K et al (2014) The relationship between total and regional corpus callosum atrophy, cognitive impairment and fatigue in multiple sclerosis patients. Mult Scler J 20:356–364
Abdul-Kareem IA, Stancak A, Parkes LM, Sluming V (2009) Regional corpus callosum morphometry: effect of field strength and pulse sequence. J Magn Reson Imaging 30:1184–1190
Haller S, Falkovskiy P, Meuli R et al (2016) Basic MR sequence parameters systematically bias automated brain volume estimation. Neuroradiology 58:1153–1160
Maubon AJ, Ferru J-M, Berger V et al (1999) Effect of field strength on MR images: comparison of the same subject at 0.5, 1.0, and 1.5 T. Radiographics 19:1057–1067
Han X, Jovicich J, Salat D et al (2006) Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. Neuroimage 32:180–194
Rutt BK, Lee DH (1996) The impact of field strength on image quality in MRI. J Magn Reson Imaging 6:57–62
Lee DH, Vellet AD, Eliasziw M et al (1995) MR imaging field strength: prospective evaluation of the diagnostic accuracy of MR for diagnosis of multiple sclerosis at 0.5 and 1.5 T. Radiology 194:257–262
Jovicich J, Marizzoni M, Sala-llonch R et al (2013) Brain morphometry reproducibility in multi-center 3 T MRI studies: a comparison of cross-sectional and longitudinal segmentations. Neuroimage 83:472–484
Despotovic I, Goossens B, Philips W (2015) MRI segmentation of the human brain: challenges, methods, and applications. Comput Math Methods Med 2015:450341
Guenette JP, Stern RA, Tripodis Y et al (2018) Automated versus manual segmentation of brain region volumes in former football players. Neuroimage Clin 18:888–896
Tardif CL, Collins DL, Pike GB (2009) Sensitivity of voxel-based morphometry analysis to choice of imaging protocol at 3 T. Neuroimage 44:827–838
Ugurbil K, Garwood M, Ellermann J et al (1993) Imaging at high magnetic fields: initial experiences at 4 T. Magn Reson Q 9:259–277
Highley JR, Esiri MM, Mcdonald B, Cortina-borja M, Herron BM, Crow TJ (1999) The size and fibre composition of the corpus callosum with respect to gender and schizophrenia: a post-mortem study. Brain 122(Pt 1):99–110
Abdul-Kareem IA, Sluming V (2008) Heschl gyrus and its included primary auditory cortex: structural MRI studies in healthy and diseased subjects. J Magn Reson Imaging 28:287–299
Aldhafeeri FM, Alghamdi J, Sluming V, Mackenzie I, Kay T (2012) Neuroanatomical correlates of tinnitus revealed by cortical thickness analysis and diffusion tensor imaging. Neuroradiology 54:883–892
FIschl B, Dale AM (2000) Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A 97:11050–11055
Mugler JP 3rd, Brookeman JR (1990) Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE). Magn Reson Med 15:152–157
Mugler JP 3rd, Brookeman JR (1991) Rapid three-dimensional T1-weighted MR imaging with the MP-RAGE sequence. J Magn Reson Imaging 1:561–567
Wang J, He L, Zheng H, Lu ZL (2014) Optimizing the magnetization-prepared rapid gradient-echo (MP-RAGE) sequence. PLoS One, 9, e96899.
Deichmann R, Schwarzbauer C, Turner R (2004) Optimisation of the 3D MDEFT sequence for anatomical brain imaging: technical implications at 1.5 and 3 T. Neuroimage 21:757–767
Thomas DL, De vita E, Deichmann R, Turner R, Ordidge RJ (2005) 3D MDEFT imaging of the human brain at 4.7 T with reduced sensitivity to radiofrequency inhomogeneity. Magn Reson Med 53:1452–1458
Gispert JD, Reig S, Pascau J, Vaquero JJ, Garcia-barreno P, Desco M (2004) Method for bias field correction of brain T1-weighted magnetic resonance images minimizing segmentation error. Hum Brain Mapp 22:133–144
Juntu J, Sijbers J, Van Dyck D, Gielen J (2005) Bias Field Correction for MRI Images. In: Kurzyński M, Puchała E, Woźniak M, żołnierek A (eds) Computer Recognition Systems. Advances in Soft Computing, vol 30. Springer, Berlin, Heidelberg
Fellner F, Holl K, Held P, Fellner C, Schmitt R, Bohm-jurkovic H (1996) A T1-weighted rapid three-dimensional gradient-echo technique (MP-RAGE) in preoperative MRI of intracranial tumours. Neuroradiology 38:199–206
Acknowledgements
The authors thank all the volunteers who participated in this study.
Funding
None.
Author information
Authors and Affiliations
Contributions
FHA has collected the data, performed the analysis and wrote the manuscript. OMA has reviewed the technical aspects, AAQ has reviewed the analysis and KMA has reviewed the final manuscript, VS has provided logistic support. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All participants received the participants’ information sheet and submitted consent forms prior to the examination. This study has been approved by the National Research Ethics Services (NRES) Committee North West-Liverpool Central.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Alhazmi, F.H., Abdulaal, O.M., Qurashi, A.A. et al. The effect of the MR pulse sequence on the regional corpus callosum morphometry. Insights Imaging 11, 17 (2020). https://doi.org/10.1186/s13244-019-0821-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13244-019-0821-8