Abstract
Background
Dirofilaria immitis, D. repens and Acanthocheilonema reconditum are the main causative agents of zoonotic canine filariosis.
Methods
We developed a combined multiplex approach for filaria and Wolbachia detection using the 28S-based pan-filarial and 16S-based pan-Wolbachia qPCRs, respectively, involving a fast typing method of positive samples using triplex qPCR targeting A. reconditum, D. immitis and D. repens, and a duplex qPCR targeting Wolbachia of D. immitis and D. repens. The approach was complemented by a duplex qPCR for the differential diagnosis of heartworms (D. immitis and Angiostrongylus vasorum) and pan-filarial cox1 and pan-Wolbachia ftsZ PCRs to identify other filarial parasites and their Wolbachia, respectively. A total of 168 canine blood and sera samples were used to validate the approach. Spearmanʼs correlation was used to assess the association between filarial species and the strain of Wolbachia. Positive samples for both the heartworm antigen-test after heating sera and at least one DNA-positive for D. immitis and its Wolbachia were considered true positive for heartworm infection. Indeed, the presence of D. repens DNA or that of its Wolbachia as well as A. reconditum DNA indicates true positive infections.
Results
The detection limit for Wolbachia and filariae qPCRs ranged from 5 × 10−1 to 1.5 × 10−4 mf/ml of blood. When tested on clinical samples, 29.2% (49/168) tested positive for filariae or Wolbachia DNA. Filarial species and Wolbachia genotypes were identified by the combined multiplex approach from all positive samples. Each species of Dirofilaria was significantly associated with a specific genotype of Wolbachia. Compared to the true positives, the approach showed excellent agreement (k = 0.98–1). Unlike D. immitis DNA, no A. vasorum DNA was detected by the duplex qPCR. The immunochromatographic test for heartworm antigen showed a substantial (k = 0.6) and a weak (k = 0.15) agreements before and after thermal pre-treatment of sera, respectively.
Conclusions
The proposed approach is a reliable tool for the exploration and diagnosis of occult and non-occult canine filariosis. The current diagnosis of heartworm disease based on antigen detection should always be confirmed by qPCR essays. Sera heat pre-treatment is not effective and strongly discouraged.

Similar content being viewed by others
Background
Canine filariosis includes diseases caused by parasitic nematodes called filariae, belonging to the order Spirurida. There are several species of veterinary and human importance. Dogs seem to be the natural hosts for several species, such as Dirofilaria immitis, D. repens, Acanthocheilonema reconditum, A. dracunculoides, Cercopithifilaria grassii, Brugia ceylonensis, B. patei, B. malayi, B. pahangi, Onchocerca lupi and Thelazia callipaeda [1,2,3,4]. These arthropod-borne filarioids produce blood, cutaneous or mucous microfilariae, where they are available to arthropod vectors [5]. The most common and medically important species affecting dogs are D. immitis, D. repens and A. reconditum [6]. In addition to their veterinary importance, they can also affect human health. Dirofilaria immitis causes pulmonary and cardiopulmonary dirofilariosis in humans and dogs, respectively. Cardiopulmonary dirofilariosis usually called heartworm disease has recently been considered as an emerging disease in Europe. In the USA, D. immitis is the most important life-threatening parasitic infection in dogs [7]. Elsewhere in the world, particularly in eastern European countries, D. repens is the most endemic parasitic nematode causing subcutaneous infection, which is less virulent but more zoonotic than that caused by D. immitis [8]. Acanthocheilonema reconditum is an occasional zoonotic agent that affects the subcutaneous tissue and the perirenal fat [9, 10] causing a common but clinically less important infection in dogs [11].
Once mature, these filarioids can produce microfilariae circulating in the bloodstream. This larval stage (L1) is also a target for the diagnosis by microscopic detection of the larvae or by detection of their DNA in the host blood [10]. Dirofilaria immitis, the agent of heartworm disease, is distributed worldwide and is responsible for heart failure in dogs after colonization of pulmonary arteries and the right ventricle, where it can be fatal if untreated. Due to the gravity of the disease, it remains the most commonly diagnosed filariosis in dogs due to the detection of antigen circulating in the blood [11,12,13]. Several problems with the current diagnostic methods have been raised, such as morphological confusion between the microfilariae of D. immitis, A. reconditum and D. repens. Commercially available diagnostic kits for the detection of D. immitis antigens may also cross-react with filarial and non-filarial nematodes, such as D. repens, A. reconditum and Onchocerca spp. [14], Spirocerca lupi and Angiostrongylus vasorum, especially the latter which can cause a cross-reaction without prior heat pre-treatment of the sera [15,16,17]. Angiostrongylus vasorum, the agent of French heartworm disease, should also be taken into account in the differential diagnosis of pulmonary disease [17]. This so-called occult heartworm is characterized by the absence of microfilaremia or an amicrofilaremia. This may result from the hostʼs immune response, low parasite load and infertility or, incidentally, the microfilaricidal effect observed in dogs receiving macrocyclic lactone prevention [14]. When the occult heartworm occurs in a co-infection with another filariosis, the diagnosis is even more challenging. The association of the heartworm with D. repens infection may result in an unexplained suppressive effect on the production of microfilariae of D. immitis [18, 19]. In such cases, the cross-reactivity between D. immitis and D. repens may result in misdiagnosis. Therefore, there is an urgent need for a more sensitive diagnostic method to detect occult as well as non-occult canine filariosis and to identify the pathogen. Detection of D. immitis has gained more and more attention; many trials have been performed for improving the quality of heartworm diagnostic tools, such as the detection of a specific antigen released by these worms [20], or the use of a recombined antigen of D. immitis for specific antibody detection [21].
The endosymbiotic intracellular bacteria of the genus Wolbachia are associated with some filarial species of two subfamilies of the Onchocercidae: Onchocercinae and Dirofilariinae [22]. These bacteria are host-specific, and each species of filarial worm is associated with a specific bacterial genotype. Wolbachia spp. have been targeted for the indirect diagnosis of D. immitis infection in dead-end hosts such as humans and cats, whereby the strong reaction of the host against the parasite prevents them to achieving their maturation, and, therefore, the production of microfilariae may not be achieved [23]. In such cases, the detection of filaria-specific Wolbachia may indicate a filarial infection and can serve as an alternative diagnostic tool in endemic areas [23, 24]. In around 40–60% of canine heartworm cases, both Wolbachia and parasite DNA may be detected using conventional PCR [25,26,27]. Indeed, the combined detection of Wolbachia and Dirofilaria DNA was suggested to improve heartworm detection [27].
In the present study, we developed a multiplex real-time PCR-based approach allowing a specific, rapid and simultaneous detection of D. immitis, D. repens and A. reconditum as well as the occult Dirofilaria spp. infections in dogs. In addition, it was completed by a duplex real-time PCR-based assay for the simultaneous detection of D. immitis and A. vasorum as a differential diagnostic for canine heartworms. The approach can be used in routinely in a diagnostic laboratory. We also evaluated the effectiveness of a novel molecular approach to conventional serological diagnosis and assessed the importance of serum heating.
Methods
Probes, primers design and PCR amplification protocol
Custom protocol and in silico validation
First, for each PCR assay, the target gene was chosen to meet the objective of each system. Fasta files were constructed from the sequences of the representative members of the family Onchocercidae or Wolbachia genotypes available in the GenBank database. The sequences were aligned using BioEdit v 7.0.5.3 software [28] to reveal the highly conserved inter- and intra-species regions as target regions for primers and probes. This region was submitted to Primer3 online software v. 0.4.0 (http://primer3.ut.ee), in order to determine valuable candidate primers and probes; the selection was based on the criteria for primer and probe design.
Physicochemical characteristics, annealing temperature and the possibility for hairpin, self- and hetero-dimers were tested using free online software Oligo-Analyzer 3.1 [29]. Primer sets and probes were also checked within DNA databases of metazoans (taxid:33208), vertebrates (taxid:7742), bacteria (taxid:2), Canidae (taxid:9608), Felidae (taxid:9682) and humans (taxid:9605) using primer-BLAST [30]. This was completed for all possible forward-reverse and probe-reverse combinations of each PCR system. Primers were synthesized by Eurogentec (Liège, Belgium) and the hydrolysis probe was synthesized by Applied BiosystemsTM (Foster City, CA, USA).
TaqMan simplex qPCR targeting filarial nematodes
The choice of the large subunit rRNA (LSU) gene, also called 28S gene, was based on several criteria such as: the tandem repetition of about 150 times in the filarial nematode genome, which improves the PCR detectability [31]; availability on GenBank for representatives of all nematode families; and sharing a highly conserved region within the Onchocercidae. The primers qFil-28S-F, qFil-28S-R and a TaqMan® hydrolysis probe (qFil-28S-P) were designed to amplify 28S gene for most filarial species (Table 1).
TaqMan triplex qPCR targeting D. immitis, D. repens and A. reconditum
The gene encoding for the cytochrome c oxidase subunit 1 gene (cox1) was selected for the development of the triplex TaqMan qPCR system targeting D. immitis, D. repens and A. reconditum (Table 1). This choice was based on the availability of cox1 for the three species on GenBank. Indeed, the cox1 gene is recognized for its high sensitivity (a high copy number relative to the nuclear gene in each cell) [32]. The cox1 gene has been described as a “barcode gene” for filarial nematodes [33]. The primers Fil.COI.749 and dg.Fil.COI.914 (Table1) were designed to amplify a 166 bp-long cox1 fragment for most members of the Onchocercidae. The system’s specificity was confined to the TaqMan probes, namely P.imm.COI.777 specific to D. immitis and P.rep.COI.871 specific to both D. repens and “Candidatus Dirofilaria (Nochtiella) honkongensis” affecting dogs and humans in Japan [34]. Finally, the probe P.rec.COI.866 is specific to A. reconditum. In the triplex TaqMan system, three different dyes were used for specific detection: FAM and VIC with a non-fluorescent quencher-TAMRA confined to D. immitis and D. repens probes, respectively; Cyanine 5 (Cy5) with a non-fluorescent quencher-BHQ-3 for the A. reconditum probe (Table 1).
TaqMan duplex qPCR targeting D. immitis and A. vasorum
The duplex cox1-based qPCR was designed (Table 1) with primers Hw.COI.723-F and Hw.COI.950-R to amplify partial cox1 gene (227 bp) of both filarial and non-filarial nematodes, including D. immitis and A. vasorum. The primers were chosen to flank the probe P.imm.COI.777, previously designed for D. immitis. In addition, we designed a new probe named A.vas.COI.813-P specific to A. vasorum. The TaqMan probes were labelled with FAM and VIC, respectively, with a non-fluorescent quencher TAMRA.
TaqMan simplex qPCR targeting Wolbachia
The 16S rDNA gene has been reported as the most commonly used gene for Wolbachia phylogeny [35]. The simplex-qPCR was developed and validated in silico for the conserved region of the first third of the 16S rDNA gene. The qPCR system (Table 1) is composed of primers Wol.16S.301f and Wol.16S.478r with the probe Wol.16S.347p targeting all Wolbachia lineages.
TaqMan duplex qPCR targeting filarial Wolbachia
Wolbachia ftsZ gene, the homologue of the eukaryotic protein tubulin, provides sufficient discrimination between Wolbachia spp. of supergroups C and D found in filarial nematodes, and those of supergroups A and B found in arthropods with a higher divergence between filarial Wolbachia of supergroups C and D [36]. The ftsZ-based duplex-qPCR was designed with primers WDiro.ftsZ.490f and wDiro.ftsZ.600r targeting filarial Wolbachia belonging to supergroup C, which includes those found in Dirofilaria sp. However, the specificity of the duplex-qPCR was confined to probes wDimm.ftsZ.523p and wDrep.ftsZ.525p specific to Wolbachia sp. of D. immitis and that of D. repens, respectively (Table 1).
Run protocols
The simplex, duplex and triplex qPCR reactions were carried out in a final volume of 20 µl, containing 5 µl of DNA template, 10 µl (2×) of Master Mix Roche (Eurogentec, Liège, Belgium). Volume of each primer per reaction was 0.5 µl (50 µM) for the simplex qPCR and 0.75 µl (50 µM) for both the duplex and triplex qPCR, with 0.5 µl of both UDG (1 U/µl) and each probe (20 µM). The final volume was made up to 20 µl using DNAse-RNAse free UltraPure water (Eurogentec, Liège, Belgium). The TaqMan cycling conditions included two hold steps at 50 °C for 2 min followed by 15 min at 95 °C, and 39 cycles of two steps at 95 °C for 30 s and 60 °C for 30 s. These reactions were performed in a CFX96 Touch Real-time PCR Detection System (Bio-Rad, Marnes-la-Coquette, France) after activating the appropriate absorption channels for the dyes used in each qPCR system.
The accumulation of the relative fluorescence units (RFUs) was recorded during the extension step of each qPCR and was used to set-up the cut-off value for each TaqMan system, according to the formula described [37]. The tolerance value was fixed at 5% for all systems. The qPCR reaction was considered positive only if the RFU value was higher than the cut-off value. qPCR data analysis was performed using the CFX Manager Software Version 3 [37].
Design of conventional PCR primers, amplification and sequencing protocols
In order to complete the molecular identification of filariae and their Wolbachia spp., we designed two sets of degenerate primers (Table 2): (i) Fspec.COI.957f and Fspec.COI.1465r targeting a 509-bp fragment of the cox1 gene of filarioids; and (ii) Wol.ftsZ.363.f and Wol.ftsZ.958.r targeting a 595-bp fragment of the ftsZ gene of Wolbachia lineages that may be associated with filarioids. All PCR reactions were carried out in a total volume of 50 µl, consisting of 25 µl (2×) of AmpliTaq Gold master mix, 18 µl of DNAse-RNAse free UltraPure water (Eurogentec, Liège, Belgium), 1 µl of each primer (20 µM) and 5 µl of DNA template (except no-template controls). The thermal cycling conditions were: incubation step at 95 °C for 15 min, followed by 40 cycles at 95 °C for 1 min, 30 s at the annealing temperature (with a different melting temperature for each PCR assay, see Table 2), 72 °C for 45 s for elongation, followed by a final extension step at 72 °C for 5 min (Table 1). PCR amplification was performed in a Peltier PTC-200 thermal cycler (MJ Research Inc., Watertown, MA, USA). Amplicons were purified using the filter plate Millipore NucleoFast 96 PCR kit following the manufacturer’s recommendations (Macherey Nagel, Düren, Germany). Purified DNA was sequenced using the BigDye® terminator v3.3 cycle sequencing kit DNA in line with the manufacturer’s instructions (Applied Biosystems). Sequencing was performed using 3500xL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) and a capillary electrophoresis fragment analyzer (Applied Biosystems). The nucleotide sequences were assembled and edited using ChromasPro 2.0.0 and were then checked using the Basic Local Alignment Search Tool (BLAST) [38].
Specificity, sensitivity and system validation
DNA samples summarized in Additional file 1: Table S1 were used for the in vitro validation of all PCR systems as follows. Nineteen samples of genomic DNA from filarial parasites were used to validate the pan-filarial 28S qPCR. DNA from eight strains of Wolbachia endosymbionts of Aedes albopictus, Anopheles gambiae, Cimex lectularius, C. hemipterus (PL13 strain), D. immitis microfilariae, D. repens, Onchocerca lupi, Wuchereria bancrofti and Brugia sp. were used for the Wolbachia 16S-based qPCR system. Dirofilaria immitis, D. repens and A. reconditum DNA were used to validate the triplex qPCR. Dirofilaria immitis and A. vasorum DNA were used to validate the duplex qPCR targeting heartworms. Finally, DNA of Wolbachia endosymbiont of D. immitis and that of D. repens were used for the duplex ftsZ-based qPCR system.
All PCR systems were tested for their specificity using several nematodes, arthropods, laboratory-maintained colonies as well as human, monkey, donkey, horse, cattle, mouse and dog DNA. DNA samples used to test the sensitivity and specificity of PCR systems are summarized in Additional file 1: Table S1 and Additional file 2: Table S2.
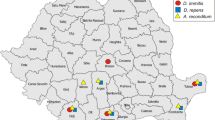
The analytical sensitivity was assessed using a 10-fold dilution of DNA templates, then standard curves and derived parameters (PCR efficiency, slope, Y-intercept and correlation coefficient) were generated using CFX Manager Software Version 3 [37]. The triplex and pan-filarial qPCR systems were challenged in detecting the related numbers of microfilariae of D. immitis, D. repens and A. reconditum. The DNA of each species was obtained from naturally infected canine blood, D. immitis (Corsica, 2018), D. repens (France, 2018) and A. reconditum (Côte d’Ivoire, 2018). First, 1 ml of each blood sample was examined by the modified Knottʼs test [9] to identify the microfilariae species and their number (Fig. 1). Then, the microfilariae concentration was adjusted to 1500 mf/ml by adding Hank’s balanced salt solution (HBSS; Gibco-BRL, Life Technologies, Eragny, France). Thereafter, two extractions were performed from 200 µl: (i) from each separately calibrated sample; and (ii) after mixing an equal volume of each of them to generate a concentration of 500 mf/ml per species. These were used to evaluate the pan-filarial and triplex qPCRs, respectively. Finally, a serial 10-fold dilution of DNA extracted from microfilaremic blood (Corsica, 2018) containing 4033 microfilariae of D. immitis was used to assess the analytical sensitivity of both the triplex and duplex (Wol-Diro ftsZ) qPCRs in the direct and the indirect detection of single infection with D. immitis.
PCR tools validation by sample screening and identification of filarial infection on biological samples
A pre-existing collection of canine blood and serum samples was used in this study. This included: (i) 8 samples composed of nematode-free laboratory Beagles from the biobank of the Veterinary Research Center of the IHU Méditerranée Infection were used as a negative control group; and (ii) 136 dogs enrolled in March 2017 from Corsica where heartworms are endemic; 7 military working dogs from France recruited on October 2018; and 17 dogs enrolled in April 2018 from Côte d’Ivoire in which blood microfilariae were recorded. Canine blood samples were collected by a veterinarian using cephalic venipuncture into a citrate and serum separator tube. The serum collected and citrate blood were then stored at − 20 °C. These 168 samples were subsequently processed for molecular and serological analysis. Genomic DNA was extracted from the blood and the microfilaria-containing tissues using the Qiagen DNA tissue extraction kit (Qiagen, Hilden, Germany) following the manufacturer’s recommendations. The extracted DNA was eluted in a total volume of 100 µl and stored at − 30 °C.
First, all DNA samples were screened for both filarial and Wolbachia DNA using the pan-filarial and pan-Wolbachia qPCRs, respectively. Then, partial cox1 and ftsZ genes were amplified and sequenced according to the previous protocol from all positive samples for filarioids and Wolbachia, respectively. Secondly, the fast typing method based on the direct identification of filarial and Wolbachia genotypes used the approach combining the triplex cox1 and duplex Wol-Diro ftsZ qPCR-based systems. Finally, all samples were screened for heartworms using the duplex cox1-based qPCR in order to differentiate between D. immitis and A. vasorum DNA.
The serological analysis was performed on all sera using the DiroCHEK® heartworm antigen test kit (Zoetis, Lyon, France). The test consisting of an enzyme-linked immunosorbent assay sandwich ELISA, targeting the antigen secreted by adult female heartworms [14]. Each serum sample was tested using two different protocols: (i) 200 µl of serum was heated at 104 °C for 10 min followed by centrifugation at 16,000×g; (ii) the second protocol was performed without heat-treatment of the sera following the recommendations by Beall et al. [39] regarding the immune complex dissociation to detect any heartworm antigen if present.
In order to evaluate the performance of molecular and serological assays in the absence of the gold standard test (necropsy), we developed the following approach to determine true positive samples. The sample was considered a true positive for heartworm if it was positive for: (i) at least one of the molecular markers of heartworm (DNA of D. immitis or its Wolbachia); and (ii) a positive antigen test after immune complex dissociation by heating sera. This approach eliminates false-positive serological results that may be obtained by increasing the detection threshold (sensitivity) after heat-treatment of the sera before use, which is subsequently confirmed by the molecular markers specific to D. immitis. Once the DNA of A. reconditum was identified, the sample was considered a true positive. Finally, samples positive for at least one DNA marker of D. repens or DNA of its Wolbachia were considered to be true positives.
Statistical analysis
Results generated through laboratory analysis were recorded in Microsoft Excel (Microsoft Corp., Redmont, USA). In order to assess how Wolbachia sp. strains correlated with filarial species, Spearmanʼs correlation coefficient was calculated. In order to evaluate the relevance of each diagnostic approach, the prevalence, correct classification, misclassification, sensitivity, specificity, false positive rate, false negative rate, positive and negative predictive value and Cohen’s Kappa (k) measure agreement was calculated. According to the scale of Landis & Koch [40], the agreement quality of Kappa values was interpreted as follows: < 0, no agreement; 0–0.2, slight agreement; 0.2–0.4, fair agreement, 0.4–0.6, moderate agreement; 0.6–0.8, substantial agreement; 0.8–1, almost perfect agreement. Statistical analyses were performed using Addinsoft 2018 (XLSTAT 2018: Data Analysis and Statistical Solution for Microsoft Excel, Paris, France).
Results
Validation of the PCR systems
The in silico validation revealed that the pan-filarial systems (28S qPCR and cox1 PCR) were specific for filarial parasites belonging to the subfamilies Dirofilariinae, Onchocercinae, Setariinae, Oswaldofilariinae, Icosiellinae and Waltonellinae. The 16S qPCR targeting Wolbachia strains was specific for all the lineages known so far. However, the ftsZ PCR showed specificity for Wolbachia strains belonging to supergroups C, D, F and J, that may be associated with filarioids. Likewise, the multiplex qPCRs were also specific for the target species without failure. For each qPCR system, primer melting temperatures were closely identical and were lower than that of the probe. Indeed, the absence of primer-dimer formation and hairpin structures was also confirmed. Furthermore, the specificity was confirmed again by an in vitro validation, as shown in Additional file 1: Table S1, where positive reaction was obtained only from the target DNA and no negative control was amplified. Despite using single-species or pooled DNAs, the specific fluorescence signals generated through the multiplex qPCR systems were successfully related to the target DNA (Additional file 3: Table S3, Additional file 4: Figure S1).
Determining assay performance characteristics
The assay characteristics were assessed for the pan-filarial, the triplex and the duplex qPCR targeting Wolbachia sp. endosymbiont of Dirofilaria spp. The analytical sensitivity of the pan-filarial qPCR was confirmed three times using D. immitis, D. repens and A. reconditum DNA sharing the same microfilariae concentration. This assay was able to detect up to 1.5 × 10−4 microfilariae per ml (mf/ml) (corresponding to 0.75 × 10−6 mf/5 μl). Efficiency ranged from 99.1 to 100.7%, with a slope from − 3.34 to − 3.30, Y-intercept values from 21.71 to 21.72 and an R2 from 0.996 to 0.999 for microfilariae of all species tested (Additional file 5: Table S4, Additional file 6: Figure S2). However, the analytical sensitivity of the triplex qPCR, using pooled DNA of three species, was confirmed by the detection of up to 5 × 10−1 mf/ml (corresponding to 2.5 × 10−3 mf/5 μl) of each species simultaneously (Additional file 7: Table S5, Additional file 8: Figure S3). qPCR efficiency ranged from 100.4 to 103.7%, with a slope from − 3.30 to − 3.24, Y-intercept values from 32.89 to 33.19 and an R2 from 0.993 to 0.999. Finally, the analytical sensitivity was confirmed for both the cox1-triplex and the ftsZ duplex qPCRs in detecting the infection by D. immitis (Additional file 9: Table S6).The detection limit was up to 4.03 × 10−2 and 4.03 × 10−1 mf/ml, respectively (corresponding to 2.01 × 10−3 and 2.01 × 10−2, respectively) mf/5 μl, qPCR efficiency was 104.8 and 100.5%, respectively, slopes were − 3.212 and − 3.309, respectively. Y-intercept values were 31.17 and 35.98, respectively, and R2 was above 0.995 for both systems (Additional file 9: Table S6, Additional file 10: Figure S4).
Molecular diagnostic approaches
Results of molecular screening followed by the sequence typing approach are detailed in Fig. 2a, b and Additional file 11: Table S7. Of the 168 samples tested, 49 (29.17%) were positive for DNA of at least one filaria species or its Wolbachia genotype. All positive results were grouped in: (i) 19 blood samples positive only for filarioid DNA; (ii) 9 samples positive only for Wolbachia DNA; and (iii) 21 samples positive for both filarial and Wolbachia DNA. Although partial cox1 and ftsZ amplicons were amplified from all positive samples for filariae and Wolbachia, cox1 gene sequence-based identification allowed the identification of the causative agent of filariosis in 35 (87.5%) out of 40 samples amplified by PCR (Fig. 2a). We here report 12 (30%) cases of D. immitis, 7 (17.5%) cases of D. repens, 15 (37.5%) cases of A. reconditum and one case of both D. repens and A. reconditum DNA. Noteworthy, the amplicon sequences were obtained separately from this latter case after serial dilution of blood before DNA extraction. However, the ftsZ gene sequence-based identification allowed the identification of Wolbachia sp. genotype in 25 (83.33%) out of 30 samples amplified by PCR (Fig. 2b). Twenty-two (73.33%) of the Wolbachia sequences were closely related to the strain identified in D. immitis (GenBank: AJ010272, 99.58%) and 3 (10%) were similar to the strain identified in D. repens (GenBank: AJ010273, 99.80%). However, sequence-based identification failed to yield sequences from amplified DNA in 5 cases for each system, which corresponds to 12.5% and 16.7% of filaria and Wolbachia DNA samples, respectively. The combined multiplex approach based on the triplex cox1 qPCR targeting filariae and the duplex qPCR targeting Wolbachia, allowed the detection of 49 samples previously considered positive based on filariae markers (Fig. 3a and Additional file 12: Table S8). The triplex cox1 qPCR identified the corresponding species from all the positive samples for filariae (n = 40, 100%). Of these, 34 (34.85%) samples had DNA from a single filarial species. Dirofilaria immitis was identified in 12 (30%), D. repens in 7 (17.5%) and A. reconditum in 15 (37.5%) of these samples. Five samples (12.5%) were positive for DNA of two filarial species, of which 4 were positive for D. immitis and D. repens and one was positive for D. repens and A. reconditum. Positivity for three filarial species was detected for only one sample (2.5%). The duplex Wol-Diro-ftsZ qPCR allowed the identification of Wolbachia genotype from all samples positive for Wolbachia DNA. Twenty-one samples (21.70%) were positive for Wolbachia sp. endosymbiont of D. immitis, three samples were positive for Wolbachia sp. endosymbiont of D. repens and five samples (16.67%) were positive for both strains. Among the 168 samples screened by duplex qPCR for heartworms (D. immitis and A. vasorum), 17 (10.12%) were positive for D. immitis DNA and no A. vasorum DNA was detected.
Venn diagram depicting the distribution of positive samples detected by molecular and serological assays. a Molecular identification of filariae and associated Wolbachia using the multiplex approach. b Comparison between molecular diagnosis and heartworm antigen detection before and after heat pre-treatment of sera
Link between Wolbachia genotype and filarial species within the infected host
The results of the distribution of filarioid markers obtained by multiplex qPCRs are shown in Fig. 3a and Additional file 12: Table S8. Interestingly, most samples which were positive for filarioid DNA were also positive for Wolbachia (21/40, 52.5%). Indeed, Wolbachia DNA was associated with at least one Dirofilaria spp. in 80% (20/25) of the samples having dirofilarial DNA. Analysis of the correlation between Wolbachia strains and Dirofilaria species are shown in Table 3. Seventy-five percent (9/12) of the samples positive for D. immitis DNA alone were also positive for Wolbachia genotype known to be associated with this filarioid, which corresponds to a significant correlation (r = 0.509, P < 0.0001). In addition, there was a significant correlation (r = 0.181, P < 0.019) between the presence of D. repens DNA alone and that of Wolbachia strain commonly associated with this filarioid. In general, the presence of D. repens DNA was correlated with the presence of DNA of both Wolbachia strains (r = 0.454, P < 0.0001). The presence of both D. immitis and D. repens DNA was associated with the presence Wolbachia endosymbiont of D. immitis (r = 0.244, P < 0.002) and also with the presence of both Wolbachia strains together (r = 0.175, P = 0.023). On the other hand, 29.63% (8/27) of the samples harboring the Wolbachia endosymbiont of D. immitis were free of filarioid DNA (Fig. 3a) and 25% (2/8) of the samples positive for Wolbachia endosymbiont of D. repens DNA were also free for filarioid DNA. No correlation was observed between A. reconditum and Wolbachia strains.
Heartworm antigen detection and infection status
Of the 168 dog sera tested for heartworm antigen, 16.67% (28) were positive before pre-treatment of the sera and were grouped into three groups (Fig. 3b): (i) 9 (5.36%) were mono-infected by heartworm and were positive for both D. immitis and its Wolbachia sp. DNA except one, which was positive for D. immitis DNA only; (ii) 10 (5.95%) were samples co-infected with at least one other filarioid detected by PCR, comprising 8 (4.76%) positive for D. repens and D. immitis and 2 (1.19%) positive for D. immitis, D. repens and A. reconditum; and (iii) 9 (5.39%) were positive for filariae other than D. immitis, with 7 (4.17%) samples positive for A. reconditum DNA only, 1 (0.6%) positive for DNA of Wolbachia endosymbiont of D. repens and 1 (0.6%) positive for DNA of both A. reconditum and Wolbachia endosymbiont of D. repens.
Once the heat pre-treatment of sera was performed, the rate of positive samples increased up to 71.43% (n = 120). Of these, 39.17% (n = 47) harbored at least one DNA marker of filarial parasites or their Wolbachia. However, two samples (1.19%), one positive for Wolbachia endosymbiont of D. immitis by qPCR and the other positive for D. repens by qPCR, remained serologically negative. For 73 samples (43.46%) no filarioid marker was detected; these were considered positive for unknown antigens (Fig. 3b and Additional file 13: Table S9). No positive results in the negative control group for both serological and molecular assays were obtained.
Performance characteristics comparison of the diagnostic tools
Once the true positive samples for each filariosis were determined, the diagnostic value was evaluated for each test in the specific detection of filariosis. The sequence typing approach combining the identification of the filariae and Wolbachia allowed the diagnosis of heartworm infection in 86.21% (25/29) of cases, which corresponds to a specificity of 82.8% and a sensitivity of 99.3%, thus resulting in an almost perfect agreement with the true positive rate (k = 0.87). Compared to the gold standard, the approach combining the multiplex qPCR systems detected one more positive sample for Wolbachia endosymbiont of D. immitis (Fig. 3b). This approach showed a sensitivity of 100% and a specificity of 99.3%, with an almost perfect agreement (k = 0.98) (Additional file 14: Table S10), whereas the detection of heartworm antigen prior to heat pre-treatment of sera showed a sensitivity of 65.5% and a specificity of 93.3%, corresponding to moderate agreement (k = 0.6). Additionally, the heat pre-treatment of sera allowed the detection of 71.4% (n = 120) including 24.17% (n = 29) positive for D. immitis infection, 15% (n= 18) positive for filariae other than D. immitis and 60.83% (n= 73) without molecular markers of filariae. The performance characteristics of this tool in detecting heartworm infection when the sera were heated were 100% sensitivity and 34.5% specificity with a slight agreement (k = 0.15) (Additional file 14: Table S10). Taking the combined multiplex approach as the gold standard, the sequence typing method had a specificity of 100% and a sensitivity of 62.6% and 94.1% for the detection of D. repens and A. reconditum, respectively. A substantial (k = 0.75) and an almost perfect (k = 0.97) agreement with the gold standard test was observed for the detection of D. repens and A. reconditum, respectively (Additional file 15: Table S11).
Discussion
qPCR system validation and assay performance characteristics
The newly developed PCR assay systems have shown specific detection of the target DNA for which they were designed. The pan-filarial 28S qPCR system aims the detection of filarial DNA from biological samples. It has been adapted for the detection of the filarial parasites known to date, such as members of the subfamilies Dirofilariinae and Onchocercinae parasitizing mammals, reptiles and birds, and those of the subfamily Setariinae, confined to large mammals, and Oswaldofilariinae parasites of reptiles, and amphibian parasites of the subfamilies Icosiellinae and Waltonellinae [5]. The LSU rRNA (28S) gene targeted by this system is known for its conserved regions between the filarial species [41]. The second qPCR system was customized for the detection of Wolbachia DNA irrespective of their lineages. It targets the first part of the 16S gene which is highly conserved between Wolbachia lineages [35]. Another qPCR system for Wolbachia targeting the 16S gene has been proposed as a complementary diagnosis from human blood of the lymphatic filariosis caused by Wuchereria bancrofti [42].
In addition to being specific, the multiplex qPCRs were discriminatory towards targeted DNA without failure (Additional file 3: Table S3). These features are directly related to the choice of the target genes, which offer sufficient discrimination between species, as is the case with the cox1 gene representing a nematode barcode [34, 43] used for the development of the triplex qPCR for D. immitis, D. repens and A. reconditum and the duplex qPCR targeting D. immitis and A. vasorum agents of heartworm diseases. Wolbachia ftsZ gene, mainly used for the characterization of Wolbachia supergroups [36], was used for the development of the duplex qPCR for both Wolbachia genotypes associated with D. immitis and D. repens. A real time PCR for Wolbachia endosymbiont of Brugia pahangi targeting the same gene has been described [44].
It is worth noting that molecular diagnosis combining the detection of filarial and Wolbachia DNA is an improvement and a tool for evaluating treatment protocols targeting filariae and Wolbachia [44]. In the present study, the analytical sensitivity of the new qPCR assays ranged from 99.3% to 107.6%, with the slope value of the standard curves ranging from − 3.34 to − 3.15 and coefficients of determination (R2) higher than 0.99. These characteristics are directly derived from the design protocol, where the formation of heterodimers and hairpins inside and between primers and probes was avoided. Primer sets share a similar melting temperature which is lower than that of the probes, offering a better sensitivity of the qPCR reaction.
The sensitivity of the pan-filarial 28S qPCR system was much higher than the triplex qPCR for the detection of D. immitis, D. repens and A. reconditum DNA, where the detection limit was 1.5 × 10−4 and 5 × 10−2 mf/ml, respectively. Indeed, the reference baseline for mitochondrial DNA retrieved from the EZ1 DNA-tissue kit was at 41.4 copies per nuclear genome [45]. Estimated genomic rRNA copy number of 150 in B. malayi [32] suggests that the 28S rRNA gene enables a high amplification efficiency rather than the mitochondrial cox1 gene of filarial nematodes. However, the sensitivity of the triplex qPCR in detecting single-species DNA of D. immitis was much higher than that of the duplex ftsZ qPCR in detecting single-species DNA of Wolbachia endosymbiont of D. immitis, where the detection limit was 4.03 × 10−3 and 4.03 × 10−1 mf/ml, respectively. Rao et al. [42], reported that filarial DNA is more frequently detected than Wolbachia DNA from W. bancrofti microfilaremic blood using qPCR assays. The difference of sensitivity could be explained by the weaker infection density by Wolbachia at this parasite larval stage [42].
Molecular diagnostic approaches
Here, we developed and assessed two molecular approaches in detecting and identifying canine filariosis. The first combined the screening and sequence typing of both filarial and Wolbachia DNA. The genomic DNA was identified with an almost perfect specificity ranging from 99.3 to 100%. However, the sensitivity ranged from moderate (62.5%) to perfect (94.1%) regarding the presence or absence of co-infection. Overlapping peaks corresponding to different nucleotides on electropherograms of the sequenced samples suggest co-infection [46]. The second approach combines two multiplex qPCR systems targeting A. reconditum, D. immitis, D. repens and the Wolbachia genotypes associated with the latter two species. All samples were positive for at least one molecular marker which were detected and identified with an almost perfect sensitivity and specificity using this approach. This method is fast, simple to use, sensitive and highly specific in detecting occult and non-occult filariosis within the infected hosts. The present results reinforce the utility of multiplex qPCR in detecting co-infections, confirm the resolution limits of the sequence typing method in the identification of co-infections [47], and avoiding the sequencing procedure needed using PCR with filaria generic primers [48].
Linkage between Wolbachia strains and filarial species within the infected host
As expected, Wolbachia DNA was significantly associated with Dirofilaria species in 80% (20/25) of the samples positive for at least one Dirofilaria spp. DNA, reinforcing the idea that this endosymbiosis relationship is present in Dirofilaria spp. and not in A. reconditum [27]. Of the samples positive for D. immitis DNA, 75% (9/12) were also found to be positive for the Wolbachia genotype known to be associated with this filarioid, resulting in a significant correlation (Table 3). As previously reported, Wolbachia DNA was detected in 64.0% of the samples positive for D. immitis [27], and in 81.6% of the samples positive for D. repens [49]. In the present study, we investigated the link between Wolbachia genotype and D. repens infection. The samples positive for a single-species DNA of D. repens had a significant correlation with the Wolbachia genotype known to be associated with this filarioid. This result corroborates the data by Vytautas et al. [49]. Interestingly, the presence of the single-species DNA of D. repens was also strongly correlated with the presence of both Wolbachia strains associated with Dirofilaria spp. This association could be explained, either by the presence of an occult co-infection with D. immitis, or by an exchange of Wolbachia strains between Dirofilaria spp. The first suggestion is supported by the fact that co-infection of D. repens and D. immitis is often associated with an occult form. This phenomenon results from a competitive suppression between microfilariae species [19]. On the other hand, Wolbachia sp. of D. immitis was detected in 29.63% (8/27) of the samples in which D. immitis DNA was not detected and, in the same samples, an antigen was detected after heat pre-treatment of sera. This result confirms the possibility to detect Wolbachia DNA in occult infections. The utility of Wolbachia as a diagnosis target for the occult heartworm disease has been demonstrated in the dead-end host, such as humans and cats, where the parasite cannot achieve its maturation and the infection might be amicrofilariaemic [23]. However, the second suggestion related to the exchange (horizontal transfer) of Wolbachia strains between Dirofilaria species is in contrast with the published data. Wolbachia transmission principally occurs via eggs of female worms (vertical transfer) [36, 50]. The vertical transfer of Wolbachia leads to the specialization of the host-symbiotic relationship [51]. Taylor et al. [52] have indicated that experimental crosses between B. pahangi and B. malayi have demonstrated Wolbachia transmission through female worms only [52]. Theoretically, exchange of Wolbachia between D. immitis and D. repens is hardly possible in natural conditions, because these filariae do not share the same site and the adult worms will not have contact inside the host organism [53]. In addition, it has been reported that each genotype of Wolbachia has a specific filarial host [36], and that live worms can release their Wolbachia endosymbionts into host tissues [52]. We believe that the presence of a specific genotype of Wolbachia is a reliable marker for the presence of its filarial host.
Performance characteristics of the heartworm antigen detection tests
In the present study, all diagnostic approaches did not react with samples from the negative control group. We assessed the diagnostic value of LISA (DiroCHEK®) in detecting heartworms. The direct exploration of heartworm antigen from sera without heating the sera showed a moderate performance, with sensitivity and specificity values of 65.52% and 93.3%, respectively. Positive antigen tests were obtained from 19 out of 29 (65.52%) of the samples determined as true positives for heartworm and these often harbored both D. immitis and its Wolbachia sp. DNA. However, 10 samples (34.38%) of which 8 (80%) harbored only DNA of the Wolbachia endosymbiont of D. immitis, remained undetected by serology. The lack of sensitivity of this assay was unexpected. This may be due in part to the presence of juvenile parasites, which do not produce detectable antigens [54]. Nevertheless, similar discordances have recently been reported, where 41 (38.7%) positive PCR-confirmed microfilaremic samples were negative for heartworm antigen [55]. Therefore, filarial Wolbachia interact with the host by activating the Th1 type protective-immune response [56], which could be implemented in the clearance of heartworm antigens.
Finally, nine out of 28 (32.14%) samples were positive for A. reconditum and/or D. repens; these filariae are known to generate a cross-reactivity in heartworm antigen tests even in the absence of heat treatment of the sera [14]. However, 29 (96.67%) of the samples positive for at least one molecular heartworm marker tested positive for heartworm antigen after heating the sera; this step has recently been added to improve the sensitivity of this test under certain conditions [15, 41]. In contrast, the heat pre-treatment of sera strongly altered the specificity of the heartworm antigen test. Cross-reactivity was observed overall in the samples positive for any filarial parasite, as well as in samples positive for unknown antigens. Filarial (D. repens and A. reconditum) and non-filarial nematodes (A. vasorum and S. lupi), are known to cross-react with heartworm antigen tests [15, 57]. Therefore, molecular and serological diagnosis of A. vasorum from canine blood showed a close efficiency [58]. Regarding the life-cycle of A. vasorum [59], the absence of its DNA from all blood samples tested in the present study is not sufficient enough to rule out infection. However, the qPCR targeting A. vasorum developed in this study can be used as a simplex for detecting parasite larvae from other biological samples, such as faeces, pharyngeal swabs as well as in the intermediate hosts. In the absence of circulating microfilaria, the antigen detection can neither confirm nor exclude the occult heartworm in area endemic for the other species that cross-react with the test, as is the case of D. repens (Fig. 2). Dirofilaria repens microfilariae induce a suppressive effect on those of D. immitis [18] which induces the occult form of the latter. The European Society of Dirofilariosis and Angiostrongylosis (ESDA) does not recommend routine heat pre-treatment of sera in an area endemic for these parasites; this is recommended for use only to resolve the discrepancy between other tests, especially when a dog is positive for the microfilaria test and negative for serology, or to confirm a suspicion of clinical disease suggestive of microfilaremia [54].
Conclusions
The molecular approach developed herein represents an improvement in the diagnosis of canine filariosis. It relies principally on TaqMan multiplex qPCR technologies. We encourage researchers to follow the molecular procedure summarized in Fig. 4. The approach allows the detection of filarial parasites as well as their Wolbachia endosymbionts at the family level from canine blood. Furthermore, we have implemented a highly sensitive and specific triplex qPCR assay for the simultaneous detection of D. immitis, D. repens and A. reconditum, the most frequent agents of canine filariosis. A duplex qPCR is presented for the simultaneous identification of Wolbachia genotypes from D. immitis and D. repens as a complementary diagnostic of canine dirofilariosis and their occult forms. Two primer sets are proposed for PCR/sequencing of filariae and Wolbachia DNAs. Finally, the approach is complemented by a duplex qPCR for D. immitis and A. vasorum, agents of canine heartworm disease. Moreover, this approach is useful in epidemiological surveillance, in diagnosis and therapeutic follow-up of both filarial parasites and their Wolbachia endosymbionts. The specific detection of Wolbachia genotypes could be used for the diagnosis of filariosis and the assessment of related pathogeny within dead-end hosts, as is the case of D. immitis infection in humans and cats. Heartworm antigen testing without heat treatment of the sera is not reliable in an endemic area with other filarial species, including A. reconditum and D. repens. We discourage the use of heat pre-treatment of sera, which significantly alters the specificity of the assay due to the cross-reactivity between many filarial and non-filarial nematodes and the possibility of false-positive results that may induce unnecessary heavy treatment of heartworm disease.
Availability of data and materials
The data supporting the conclusions of this article are included within the article and its additional files.
Abbreviations
- FAM:
-
6-FAM (6-carboxyfluorescein)
- VIC:
-
VIC (2′-chloro-phenyl-1,4- dichloro-6-carboxyfluorescein)
- Cy5:
-
cyanine 5
- TAMRA:
-
tetra-methyl-rhodamine
- BHQ:
-
black hole quencher
References
Ravindran R, Varghese S, Nair SN, Balan VM, Lakshmanan B, Ashruf RM, et al. Canine filarial infections in a human Brugia malayi endemic area of India. Biomed Res Int. 2014;2014:630160.
Latrofa MS, Annoscia G, Colella V, Cavalera MA, Maia C, Martin C, et al. A real-time PCR tool for the surveillance of zoonotic Onchocerca lupi in dogs, cats and potential vectors. PLoS Negl Trop Dis. 2018;12:e0006402.
Seixas F, Travassos P, Coutinho T, Lopes AP, Latrofa MS, dos Pires MA, et al. The eyeworm Thelazia callipaeda in Portugal: current status of infection in pets and wild mammals and case report in a beech marten (Martes foina). Vet Parasitol. 2018;252:163–6.
Otranto D, Dantas-Torres F, Brianti E, Traversa D, Petrić D, Genchi C, et al. Vector-borne helminths of dogs and humans in Europe. Parasit Vectors. 2013;6:16.
Adamson RC. Nematode parasites of vertebrates. Their development and transmission. Wallingford: CAB International; 2000.
Ilie MS, Imre K, Hotea I, Dărăbuș G. Survey of canine dirofilariosis from south-western Romania—preliminary results. In: 3rd European Dirofilaria Days, 21–22 June 2012, Parma, Italy; 2012. p. 68.
Simón F, González-Miguel J, Diosdado A, Gómez PJ, Morchón R, Kartashev V. The complexity of zoonotic filariasis episystem and its consequences: a multidisciplinary view. Biomed Res Int. 2017;2017:6436130.
Genchi C, Kramer L. Subcutaneous dirofilariosis (Dirofilaria repens): an infection spreading throughout the old world. Parasit Vectors. 2017;10:517.
Magnis J, Lorentz S, Guardone L, Grimm F, Magi M, Naucke TJ, et al. Morphometric analyses of canine blood microfilariae isolated by the Knott’s test enables Dirofilaria immitis and D. repens species-specific and Acanthocheilonema (syn. Dipetalonema) genus-specific diagnosis. Parasit Vectors. 2013;6:48.
Nutman BT. Filarial infections. In: Jong EC, Sanford CA, editors. The travel and tropical medicine manual. 4th ed. Philadelphia: Saunders/Elsevier; 2008. p. 611–25.
Luca F, Luigi V. Filarial infections. In: Day MJ, editor. Arthropod-borne infectious diseases of the dog and cat. 2nd ed. Boca Raton: CRC Press; 2016.
Simsek S, Ciftci AT. Serological and molecular detection of Dirofilaria species in stray dogs and investigation of Wolbachia DNA by PCR in Turkey. J Arthropod Borne Dis. 2016;10:445–53.
Bowman DD, Atkins CE. Heartworm biology, treatment, and control. Vet Clin North Am Small Anim Pract. 2009;39:1127–58.
Little S, Saleh M, Wohltjen M, Nagamori Y. Prime detection of Dirofilaria immitis: understanding the influence of blocked antigen on heartworm test performance. Parasit Vectors. 2018;11:186.
Aroch I, Rojas A, Slon P, Lavy E, Segev G, Baneth G. Serological cross-reactivity of three commercial in-house immunoassays for detection of Dirofilaria immitis antigens with Spirocerca lupi in dogs with benign esophageal spirocercosis. Vet Parasitol. 2015;211:303–5.
Venco L, Manzocchi S, Genchi M, Kramer LH. Heat treatment and false-positive heartworm antigen testing in ex vivo parasites and dogs naturally infected by Dirofilaria repens and Angiostrongylus vasorum. Parasit Vectors. 2017;10:476.
Rinaldi L, Cortese L, Meomartino L, Pagano TB, Pepe P, Cringoli G, et al. Angiostrongylus vasorum: epidemiological, clinical and histopathological insights. BMC Vet Res. 2014;10:236.
Monica A, Matei IA, Amico GD, Bel LV, Dumitrache MO, Modrý D, et al. Dirofilaria immitis and D. repens show circadian co-periodicity in naturally co-infected dogs. Parasit Vectors. 2017;10:116.
Mircean M, Ionică AM, Mircean V, Györke A, Codea AR, Tăbăran FA, et al. Clinical and pathological effects of Dirofilaria repens and Dirofilaria immitis in a dog with a natural co-infection. Parasitol Int. 2017;66:331–4.
Sassi AJ, Geary JF, Leroux LP, Moorhead AR, Satti M, Mackenzie CD, et al. Identification of Dirofilaria immitis proteins recognized by antibodies from infected dogs. J Parasitol. 2014;100:364–7.
Solgi R, Sadjjadi SM, Mohebali M, Zarei Z, Golkar M, Raz A. Development of new recombinant DgK antigen for diagnosis of Dirofilaria immitis infections in dogs using ELISA technique and its comparison to molecular methods. Iran Biomed J. 2018;22:283–9.
Martin C, Gavotte L. The bacteria Wolbachia in filariae, a biological Russian dolls’system: new trends in antifilarial treatments. Parasite. 2010;17:79–89.
Turb ME, Zambon E, Zannoni A, Russo S, Gentilini F. Detection of Wolbachia DNA in blood for diagnosing filaria-associated syndromes in cats. J Clin Microbiol. 2012;50:2624–30.
Dingman P, Levy JK, Kramer LH, Johnson CM, Lappin MR, Greiner EC, et al. Association of Wolbachia with heartworm disease in cats and dogs. Vet Parasitol. 2010;170:50–60.
Landum M, Ferreira CC, Calado M, Alho AM, Maurício IL, Meireles JS, et al. Detection of Wolbachia in Dirofilaria infected dogs in Portugal. Vet Parasitol. 2014;204:407–10.
Rossi MID, Aguiar-Alves F, Santos S, Paiva J, Bendas A, Fernandes O, et al. Detection of Wolbachia DNA in blood from dogs infected with Dirofilaria immitis. Exp Parasitol. 2010;126:270–2.
Maia C, Altet L, Serrano L, Cristóvão JM, Tabar MD, Francino O, et al. Molecular detection of Leishmania infantum, filariae and Wolbachia spp. in dogs from southern Portugal. Parasit Vectors. 2016;9:170.
Hall T, Biosciences I, Carlsbad C. BioEdit: an important software for molecular biology. GERF Bull Biosci. 2011;2:60–1.
Owczarzy R, Tataurov AV, Wu Y, Manthey JA, McQuisten KA, Almabrazi HG, et al. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36:163–9.
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134.
Bik HM, Fournier D, Sung W, Bergeron RD, Thomas WK. Intra-genomic variation in the ribosomal repeats of nematodes. PLoS ONE. 2013;8:e78230.
Lagatie O, Merino M, Debrah LB, Debrah AY, Stuyver LJ. An isothermal DNA amplification method for detection of Onchocerca volvulus infection in skin biopsies. Parasit Vectors. 2016;9:624.
Ferri E, Barbuto M, Bain O, Galimberti A, Uni S, Guerrero R, et al. Integrated taxonomy: traditional approach and DNA barcoding for the identification of filarioid worms and related parasites (Nematoda). Front Zool. 2009;6:1.
Yilmaz E, Fritzenwanker M, Pantchev N, Lendner M, Wongkamchai S, Otranto D, et al. The mitochondrial genomes of the zoonotic canine filarial parasites Dirofilaria (Nochtiella) repens and Candidatus Dirofilaria (Nochtiella) honkongensis provide evidence for presence of cryptic species. PLoS Negl Trop Dis. 2016;10:e0005028.
Willems A, Watteyne S, Mertens J, Gillis M, Vandekerckhove TTM, Swing JG. Phylogenetic analysis of the 16S rDNA of the cytoplasmic bacterium Wolbachia from the novel host Folsomia candida (Hexapoda, Collembola) and its implications for wolbachial taxonomy. Fems Microbiol Lett. 1999;180:279–86.
Bandi C, Anderson TJC, Genchi C, Blaxter ML. Phylogeny of Wolbachia in filarial nematodes. Proc R Soc B Biol Sci. 1998;265:2407–13.
Bio‐Rad, L. Real-time PCR applications guide. Hercules: Bio-Rad Laboratories Inc; 2006. p. 41.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Beall MJ, Arguello-Marin A, Drexel J, Liu J, Chandrashekar R, Alleman AR. Validation of immune complex dissociation methods for use with heartworm antigen tests. Parasit Vectors. 2017;10:481.
Landis JR, Koch GG. An application of hierarchical kappa-type statistics in the assessment of majority agreement among multiple observers. Biometrics. 1977;33:363–74.
Chilton NB, Huby-Chilton F, Gasser RB. First complete large subunit ribosomal RNA sequence and secondary structure for a parasitic nematode: phylogenetic and diagnostic implications. Mol Cell Probes. 2003;17:33–9.
Rao RU, Atkinson LJ, Ramzy RM, Helmy H, Farid HA, Bockarie MJ, et al. A real-time PCR-based assay for detection of Wuchereria bancrofti DNA in blood and mosquitoes. Am J Trop Med Hyg. 2006;74:826–32.
Prosser SWJ, Velarde-Aguilar MG, León-Règagnon V, Hebert PDN. Advancing nematode barcoding: a primer cocktail for the cytochrome c oxidase subunit I gene from vertebrate parasitic nematodes. Mol Ecol Resour. 2013;13:1108–15.
Simoncini L, Casiraghi M, Bazzocchi C, Sacchi L, Bandi C, Genchi C. Real-time PCR for quantification of the bacterial endosymbionts (Wolbachia) of filarial nematodes. Parassitologia. 2001;43:173–8.
Fazzini F, Schöpf B, Blatzer M, Coassin S, Hicks AA, Kronenberg F, et al. Plasmid-normalized quantification of relative mitochondrial DNA copy number. Sci Rep. 2018;8:15347.
Kommedal Ø, Karlsen B, Sæbø Ø. Analysis of mixed sequencing chromatograms and its application in direct 16S rRNA gene sequencing of polymicrobial samples. J Clin Microbiol. 2008;46:3766–71.
Irshad M, Gupta P, Mankotia DS, Ansari MA. Multiplex qPCR for serodetection and serotyping of hepatitis viruses: a brief review. World J Gastroenterol. 2016;22:4824–34.
Rishniw M, Barr SC, Simpson KW, Frongillo MF, Franz M, Dominguez Alpizar JL. Discrimination between six species of canine microfilariae by a single polymerase chain reaction. Vet Parasitol. 2006;135:303–14.
Sabūnas V, Radzijevskaja J, Sakalauskas P, Petkevičius S, Karvelienė B, Žiliukienė J, et al. Dirofilaria repens in dogs and humans in Lithuania. Parasit Vectors. 2019;12:177.
Rao RU. Endosymbiotic Wolbachia of parasitic filarial nematodes as drug targets. Indian J Med Res. 2005;122:199–204.
Taylor MJ, Bandi C, Hoerauf A. Wolbachia bacterial endosymbionts of filarial nematodes. Adv Parasitol. 2005;60:245–84.
Taylor MJ, Hoerauf A. Wolbachia bacteria of filarial nematodes. Parasitol Today. 1999;15:437–42.
McCall JW, Genchi C, Kramer LH, Guerrero J, Venco L. Heartworm disease in animals and humans. Adv Parasitol. 2008;66:193–285.
Nelson CT, McCall JW, Jones S, Moorhead A. Current canine guidelines for the prevention, diagnosis, and management of heartworm (Dirofilaria immitis) infection in dogs. American Heartworm Society; 2018. https://www.heartwormsociety.org/veterinary-resources/american-heartworm-society-guidelines. Accesed 25 Dec 2019.
Alho AM, Landum M, Ferreira C, Meireles J, Gonçalves L, de Carvalho LM, et al. Prevalence and seasonal variations of canine dirofilariosis in Portugal. Vet Parasitol. 2014;206:99–105.
Tabar MD, Altet L, Martínez V, Roura X, Veterinario H, Vicente S, et al. Wolbachia, filariae and Leishmania coinfection in dogs from a Mediterranean area. J Small Anim Pract. 2013;54:174–8.
Schnyder M, Deplazes P. Cross-reactions of sera from dogs infected with Angiostrongylus vasorum in commercially available Dirofilaria immitis test kits. Parasit Vectors. 2012;5:258.
Jefferies R, Morgan ER, Helm J, Robinson M, Shaw SE. Improved detection of canine Angiostrongylus vasorum infection using real-time PCR and indirect ELISA. Parasitol Res. 2011;109:1577–83.
Ferdushy T, Hasan MT. Angiostrongylus vasorum: the “French heartworm”. Parasitol Res. 2010;107:765–71.
Acknowledgements
We gratefully thank Sebastien Cortaredona for assistance with statistical analysis. We also thank Professor Domenico Otranto who provided us with positive control DNA. We express our gratitude to Zoetis who provided us with the DiroCHEK® tests.
Funding
This study was supported by the Institut-Hospitalo-Universitaire (IHU) Méditerranée Infection, the National Research Agency under the program “Investissements d’avenir”, reference ANR-10-IAHU-03, the Région Provence-Alpes-Côte d’Azur and European funding FEDER PRIMI. Funding sources had no role in the design or conduct of the study; collection, management, analysis or interpretation of the data; or preparation, review or approval of the manuscript.
Author information
Authors and Affiliations
Contributions
YL, BD, MV and OM designed the study. YL, BD, OM designed, and YL carried out the data analysis. YL, MV, EAN and OM drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
DNA used as a positive control in this study.
Additional file 2: Table S2.
DNA used as a negative control in this study.
Additional file 3: Table S3.
In vitro validation of the triplex cox1-based qPCR.
Additional file 4: Figure S1.
Assessment of the specificity of the triplex cox1-based qPCR in detecting the target DNA.
Additional file 5: Table S4.
Analytical sensitivity of the pan-filarial 28S-based qPCR in detecting single-species DNA.
Additional file 6: Figure S2. a
, b, c-1. Efficiency of pan-filarial 28S-based qPCR using single-species DNA of D. immitis, D. repens and A. reconditum. a, b, c-2. Standard curves generated from serial 10-fold dilution of each DNA.
Additional file 7: Table S5.
Analytical sensitivity of the triplex cox1-based qPCR using pooled DNA.
Additional file 8: Figure S3. a
Efficiency of the triplex cox1-based qPCR using pooled DNA. b Standard curves generated from serial 10-fold dilution of DNA.
Additional file 9: Table S6.
Analytical sensitivity of the triplex cox1 and the duplex ftsZ-based qPCRs in detecting single-species DNA of D. immitis and its Wolbachia.
Additional file 10: Figure S4. a, b
Efficiency of the triplex cox1 and the duplex ftsZ-based qPCRs using single-species DNA of D. immitis and its Wolbachia. c, d Standard curves generated from serial 10-fold dilution of DNA.
Additional file 11: Table S7.
Samples distribution according to filariae and Wolbachia DNA detected by the sequence typing approach.
Additional file 12: Table S8.
Samples distribution according to filariae and Wolbachia DNA detected by the multiplex approach.
Additional file 13: Table S9.
Samples tested positive by at least by one diagnostic approach.
Additional file 14: Table S10.
Performance of molecular and serological assays in detecting D. immitis infection.
Additional file 15: Table S11.
Performance of molecular approaches in detecting D. repens and A. reconditum.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Laidoudi, Y., Davoust, B., Varloud, M. et al. Development of a multiplex qPCR-based approach for the diagnosis of Dirofilaria immitis, D. repens and Acanthocheilonema reconditum. Parasites Vectors 13, 319 (2020). https://doi.org/10.1186/s13071-020-04185-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04185-0