Abstract
Background
Flea-borne spotted fever is a zoonosis caused by Rickettsia felis, a Gram-negative obligate intracellular bacterium. The disease has a worldwide distribution including western and eastern sub-Saharan Africa where it is associated with febrile illness in humans. However, epidemiology and the public health risks it poses remain neglected especially in developing countries including Zambia. While Ctenocephalides felis (cat fleas) has been suggested to be the main vector, other arthropods including mosquitoes have been implicated in transmission and maintenance of the pathogen; however, their role in the epidemiological cycle remains to be elucidated. Thus, the aim of this study was to detect and characterize R. felis from animal hosts and blood-sucking arthropod vectors in Zambia.
Methods
Dog blood and rodent tissue samples as well as cat fleas and mosquitoes were collected from various areas in Zambia. DNA was extracted and screened by polymerase chain reaction (PCR) targeting genus Rickettsia and amplicons subjected to sequence analysis. Positive samples were further subjected to R. felis-specific real-time quantitative polymerase chain reactions.
Results
Rickettsia felis was detected in 4.7% (7/150) of dog blood samples and in 11.3% (12/106) of rodent tissue samples tested by PCR; this species was also detected in 3.7% (2/53) of cat fleas infesting dogs, co-infected with Rickettsia asembonensis. Furthermore, 37.7% (20/53) of cat flea samples tested positive for R. asembonensis, a member of spotted fever group rickettsiae of unknown pathogenicity. All the mosquitoes tested (n = 190 pools) were negative for Rickettsia spp.
Conclusions
These observations suggest that R. felis is circulating among domestic dogs and cat fleas as well as rodents in Zambia, posing a potential public health risk to humans. This is because R. felis, a known human pathogen is present in hosts and vectors sharing habitat with humans.
Similar content being viewed by others
Background
Rickettsia felis is an obligate intracellular gram-negative alpha-proteobacterium that causes zoonotic cat flea typhus also known as flea-borne spotted fever in humans. The bacterium was first described by Adams et al. in 1990 in the cytoplasm of midgut cells of the cat flea Ctenocephalides felis [1]. It was later characterized as R. felis and confirmed as the causative agent of flea-borne spotted fever in humans [2, 3]. The genus Rickettsia is divided into three groups based on genotyping: the ancestral group, the typhus group and the spotted fever group to which R. felis belongs [4].
Spotted fever rickettsioses are one of the emerging infectious diseases to which little attention has been accorded. The first human case of R. felis infection was reported in 1994 in Texas, USA [3]. Since then, it remained neglected until its emergence as a cause of febrile illness in sub-Saharan Africa [5]. It has since been reported in eastern and western sub-Saharan Africa [6, 7] and North Africa [8]. Rickettsia felis infections in humans have also been reported in the USA, Brazil and Mexico in the Americas with varying clinical signs and severity, in Taiwan, Thailand, South Korea, France, Germany, Spain and other parts of the world [8, 9]. This worldwide distribution of R. felis infections in humans resulted into its consideration as a global emerging threat to human health [9, 10]. Once a susceptible human is infected, rickettsiae mainly invade endothelial cells and cause vasculitis, acute flu-like symptoms, fever, chills, headache, skin rash and photophobia [11] as the clinical manifestations of flea-borne spotted fever.
Cat fleas have been considered as the hosts and vectors of R. felis [8]. However, recent reports have suggested other arthropods such as other flea species [12], ticks [13], mites [14], and booklice [15] as potential vectors in the epidemiological cycle through detection of R. felis using molecular techniques. Rickettsia felis has also been detected in Aedes albopictus in Gabon [16] and was experimentally proven to be transmitted by Anopheles gambiae in a mouse model [17]. This claim was further strengthened by the elucidation of co-infection with malaria and R. felis among febrile patients in malaria-endemic areas of Senegal, suggesting co-transmission by mosquitoes [7].
Domestic dogs and cats are considered mammalian reservoir hosts for R. felis [18, 19]. Other proposed mammalian reservoirs include domestic/peridomestic rodents, opossums and raccoons which are documented to maintain the pathogen in peridomestic areas [20, 21]. However, what is unclear is the interaction between the domestic/peridomestic cycles for pathogen transmission and maintenance in nature. Cat fleas may play an essential role in both transmission cycles resulting in potential risk of human exposure due to their indiscriminate feeding behavior [22]. Additionally, R. felis can be circulating in cat fleas via transstadial and transovarial transmission consequently fleas acting as reservoirs and vectors. Therefore, understanding the host-vector relationship and epidemiology of R. felis is vital for pathogen surveillance and effective control measures in case of outbreaks as these animals share habitat with humans.
Although R. felis is frequently detected in western and eastern sub-Saharan Africa, little is known about its epidemiology and the public health risk it poses in Zambia. On the other hand, other spotted fever group rickettsiae have been reported from Zambia. Tamaki et al. [23] reported 16.7% seroprevalence for antibodies against Rickettsia conorii in humans despite cross-reactivity of spotted fever group rickettsia and R. felis on serology having been documented [24], hence possibility of misdiagnosing some closely related species cannot be excluded. Later Nakayima et al. [25] found molecular evidence of spotted fever group rickettsiae closely related to Rickettsia africae (the causative agent for African tick bite fever) in free-ranging non-human primates in Mambwe District near Luangwa National Park, Zambia. With the endemic cases of malaria [26] little attention has been given to other pathogens that cause febrile illness, hence possibilities of misdiagnosis could be high. A study in Kenya found strong evidence of the association of R. felis with a fever of unknown origin [27].
To date, no studies have been carried out to elucidate the prevalence of R. felis in animals, humans and arthropods in Zambia. Hence, it is of prime importance to understand the epidemiology of R. felis both in mammalian hosts and arthropod vectors/potential vectors as well as the risk of human transmission. The aim of this study was thus to detect and characterize R. felis from animal hosts and possible arthropod vectors in Zambia. The findings would provide the baseline for future surveillance of Rickettsia spp. in Zambia and risk assessment of human exposure to the infection.
Methods
Sample collection
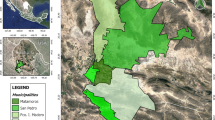
Blood samples from visibly healthy domestic dogs were collected from Lusaka District in Lusaka Province, and from Mazabuka and Monze districts in the Southern Province of Zambia (Fig. 1) in 2015. Genomic DNA was extracted from blood using the DNAzol kit (Molecular Research Center, Cincinnati, OH) according to manufacturers’ protocols. Genomic DNA samples (n = 106) from rodents were collected in a previous study [28]. The rodent species were identified by partial sequence of the cytochrome c oxidase subunit 1 gene (cox1) or cytochrome b gene (cytb). The analyzed species included Mastomys sp., Steatomys sp., Lemniscomys sp., Saccostomus sp., Rattus rattus and the gerbil.
Cat flea (Ctenocephalides felis) samples (n = 53) infesting different dogs in Mazabuka District, Southern Province, were collected in 2016 and identified to species level using morphological characteristics [29]. After identification to the species level, each cat flea was separately homogenized and DNA was extracted. Mosquitoes of different species were collected by using the CDC and BG traps in Zambia during 2016–2017. They were then identified to the species level using morphological taxonomic keys. The mosquitoes were then pooled (n ≤ 10) based on sex and species, homogenized and DNA extracted using Takara Simpleprep kit (Takara, Shiga, Japan).
PCR amplification and amplicon sequencing
Conventional PCR targeting the partial gltA gene for genus Rickettsia was essentially performed using Ampdirect plus buffer (Shimadzu, Tokyo, Japan) with BioTaq Polymerase (Bioline, London, UK) as described by manufacturers. The PCR was performed at 94 °C DNA denaturation, 54 °C annealing and 72 °C extension temperatures for 35 cycles using the primers described by Gaowa et al. [30]. The amplicons were resolved on a 1.2% agarose gel stained with GelRed (Biotium, Hayward, CA) and visualized under UV light. Positive samples with gltA PCR were processed for PCR targeting the outer membrane protein A (OmpA) and outer membrane protein B (OmpB) genes using primers designed in this study. The OmpA and OmpB gene fragment amplification primers were designed using the Primer-blast tool in NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The OmpA and OmpB gene sequences for R. felis accession number CP000053 retrieved from GenBank were used as references for primer design. The PCRs were essentially conducted as described above with modification of annealing temperature (48 °C for the OmpA and OmpB) and amplicons were electrophoresed on 2% and 1.2% agarose gels respectively. Rickettsia monacensis DNA extracted from cell culture was used as a positive control to avoid sample contamination by R. felis DNA from the positive control and nuclease-free water was used as a negative control in all assays. All samples that were gltA PCR-positive were further subjected to partially R. felis-specific qPCR targeting biotin synthase gene (bioB) and a R. felis unique segment of the outer membrane protein gene (RfelB) using primers and probe described previously [31, 32]. All primers and probes used are shown in Table 1.
PCR products were purified using ExoSap-IT (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. DNA concentration was measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific). Amplicons were cycle-sequenced by BigDye Terminator v3.1 cycle sequencing kit (Thermo Fisher Scientific) with the same primers as used for PCR amplification and analyzed on a 3500 × l Genetic Analyzer (Applied Biosystems, Foster City, CA). The DNA sequences obtained were submitted to the DNA Data Bank of Japan (DDBJ) (http://www.ddbj.nig.ac.jp) under the accession numbers LC431490–LC431502.
Alignment and phylogenetic analysis
The newly generated nucleotide sequences were assembled and aligned with relevant sequences from GenBank in NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) using ClustalW aligner [33]. Phylogenetic analysis of sequences obtained in the present study along with reference sequences was performed by Neighbor-Joining method. Bootstrap support was based on 500 replicates. Alignment and phylogenetic analyses were conducted in MEGA 7.0.2.1 software [34].
Statistics
Statistical analysis was performed by the Chi-square statistical test to determine the statistical difference in Rickettsia sp. prevalence among dogs from densely populated urban areas of Lusaka District and sparsely populated rural areas of Monze and Mazabuka Districts. Differences were considered significant at P ≤ 0.05.
Results
A sample was considered positive to the species level upon being positive for at least two markers (Additional file 1: Table S1). Based on this criterion, a sample was considered R. felis-positive when the partial gltA gene fragment nucleotide have homology of more than 99% to R. felis strain URRXCal2 (GenBank: CP000053) reference sequence [4] and, partially R. felis-specific bioB or R. felis unique fragment of outer membrane protein B (RfelB) qPCR was positive. Furthermore, samples positive for both qPCR markers were considered to be positive for R. felis. Sequence homology based on outer membrane protein A (OmpA) and outer membrane protein B (OmpB) genes was used to further confirm other Rickettsia species including Rickettsia felis-like organisms (RFLOs) to the species level. Isolates not meeting above criteria were identified to the spotted fever group level.
Rickettsia felis was detected in 4.7% (7/150) of the dogs from Lusaka and Mazabuka, and 3.7% (2/53) of the cat fleas collected from Mazabuka. Furthermore, 11.3% (12/106) of the rodents from Lusaka and Namwala were also positive for R. felis (Table 2). All the positive rodents were of the Mastomys sp. The obtained sequences of gltA gene fragment from dog and rodent samples were 99–100% similar to R. felis strain URRXCal2 (GenBank: CP000053) and further confirmed by R. felis specific qPCR (Additional file 1: Table S1). All dog blood samples from Monze (n = 50) were negative for Rickettsia spp.
Among the analyzed cat flea samples obtained from Mazabuka, 3.7% (2/53) were positive for R. felis-specific RfelB qPCR (Table 2, Additional file 1: Table S1). Interestingly, their gltA sequences showed higher homology to R. asembonensis than R. felis suggesting that these cat fleas were co-infected with R. felis and R. asembonensis. The qPCRs targeting bioB (mean Ct = 26) which is specific for both R. felis and R. asembonensis were also positive, but their Ct values were lower than those for RfelB (mean Ct = 36). The lower Ct values of bioB qPCR as compared to RfelB suggests that the population of R. asembonensis was higher than that of R. felis in the cat flea samples. It is also consistent with the observed R. asembonensis type gltA sequences. The presence of R. asembonensis was further confirmed by sequence homology based on the OmpA and OmpB gene fragments. The sequences of the gltA, OmpA and OmpB gene fragments obtained from cat fleas were 100%, 99% and 100% similar to Rickettsia asembonensis sequences with accession numbers JN315968, KY650698 and KY650699, respectively. The overall positive rate for R. asembonensis in Mazabuka cat fleas was 37.7% (20/53).
As shown in Fig. 2, the gltA sequences from dog blood and rodent tissue samples clustered with R. felis in the phylogenetic analysis regardless of the sampling location. In contrast, those of cat flea samples were separated from R. felis sequences and clustered with R. asembonensis. The gltA gene sequences obtained from rodent samples were more diversified from the reference sequences previously identified in cat fleas (Fig. 2). Further phylogenetic analysis based on the rOmpB gene fragment from cat flea samples similarly clustered with R. asembonensis (data not shown).
Unrooted phylogenetic tree based on sequences of the gltA gene. Neighbor-joining phylogenetic relationships of spotted fever group rickettsiae detected in cat fleas, dogs and rodents in Zambia. The tree was based on 321 bp fragment of gltA gene. Bold labels represent sequences from this study with the detected species name, accession number, and host and place of collection in the label
One hundred ninety of pooled samples from different mosquito species, Culex spp., Aedes spp. and Anopheles spp. (listed in Additional file 1: Table S2) collected from Lusaka, Mpulungu and Isoka districts were all negative for Rickettsia spp. by gltA PCR. Among the analyzed mosquito samples, 81.1% (154/190) were collected from Lusaka District, the same location where R. felis was detected in dogs and rodents. Among the sampled mosquitoes from Lusaka District, 87% (134/154) belonged to Culex spp. with the others being Anopheles spp. and Aedes spp. In contrast to Lusaka District, 87.8% (29/33) of the samples from Mpulungu District were Aedes spp. (Additional file 1: Table S2) suggesting that Culex spp. are more predominant in the urban Lusaka township.
Discussion
Investigation of neglected zoonotic pathogens in domestic pets and vectors infesting them is important in the control and eradication of zoonotic diseases. In this study, we found 4.7% (7/150) prevalence of R. felis in domestic dogs. The detected genotype is identical to reported one found in GenBank which was isolated from cat fleas and mosquitoes, and is known to cause flea-borne spotted fever in humans [5, 35]. This highlights the significance of domestic dogs as potential reservoirs of R. felis. It is therefore important for dog owners to be aware of rickettsia infection risk as concurrent R. felis infections in a positive dog and its infected owners have already been reported [36].
Among the blood samples from dogs in Lusaka, Mazabuka and Monze districts, it was found that dogs from the urban areas of Lusaka have a significantly higher prevalence of Rickettsia sp. (40%, 20/50) than dogs from the rural areas of Mazabuka (10%, 5/50) and Monze (0%, 0/50). This could be due to the off-host nature of fleas resulting in feeding on many animals in close proximity. The high dog population density and closeness of households in the urban areas of Zambia could possibly make it easy for cat fleas to feed on many dogs within a close proximity. Therefore, the host diversity of cat fleas [37] in different demographical setups could help explain the differences in prevalences of Rickettsia sp. in dogs from rural and urban areas.
It is known that the cat flea is a vector for flea-borne spotted fever causative agent. In this study, R. felis was also detected in cat fleas in the same sampling place (Mazabuka District) where dogs were positive for R. felis. This molecular-based evidence implies that R. felis is circulating with domestic dogs serving as reservoirs and cat fleas as vectors in Zambia. This is of public health significance as the bacterium is potentially maintained among domestic dogs and cat fleas as earlier reported [38]. This results in a potential risk of human infections through cat flea bites as they feed indiscriminately, hence effective vector-borne infection control measures are necessary to prevent zoonotic disease outbreaks.
High prevalence of R. asembonensis (41.5%) was also observed with low prevalence of R. felis (3.7%) in cat fleas. This result is in agreement with previous studies reporting up to 100% prevalence of R. asembonensis in cat fleas with low R. felis prevalence [37, 39,40,41,42,43,44]. Interestingly, two of the cat flea samples positive for R. felis were also positive for R. asembonensis. These findings provide evidence for co-infection of R. felis and R. asembonensis in cat fleas. Furthermore, it is important to investigate the pathogenicity of R. asembonensis to understand its infectivity in dogs and more importantly humans [45, 47]. This is because cat fleas feed indiscriminately, hence the potential risk of human exposure through flea bites.
This study also presents molecular evidence of R. felis in rodents Mastomys sp. in Zambia, suggesting that the latter could be potential reservoirs of R. felis. Rodents have been implicated as reservoir hosts for diverse zoonotic pathogens ranging from viruses, protozoans, fungi, helminths to bacteria [46] including Rickettsia spp. [20]. Limited studies have been conducted to understand the role of rodents in the epidemiology and ecology of R. felis [20, 42, 47, 48], despite many studies having detected R. felis DNA in fleas infesting rodents [8, 42, 43, 49]. This result adds Mastomys sp. to the rodent species implicated to be potentially maintaining R. felis in nature. Rodents are suggested to be vital for maintenance of R. felis resulting in sporadic cases of rickettsioses in humans as they share habitat with humans and dogs in the presence of fleas as vectors [50]. Therefore, interplay among mammalian hosts, fleas and humans need to be clarified in order to understand the zoonosis transmission cycle and possible control measures.
Many other blood-sucking arthropods with the potential to transmit rickettsiae are present in Zambia. Therefore, surveillance for neglected vector-borne zoonoses such as spotted fever rickettsioses posing a potential emerging public health threat is necessary. It is for this reason that different mosquito species, including Culex spp., Aedes spp. and Anopheles spp. (Additional file 1: Table S2) were screened for Rickettsia spp. and were notably all negative. This suggests that mosquitos may not be playing an essential role in the epidemiology of R. felis in Zambia despite earlier reports [7, 35]. The role of these mosquito species in R. felis epidemiology is still unclear despite recent advances in transmission potential studies, hence further studies are needed [7, 16, 17].
Conclusions
This study provides, to our knowledge for the first time in Zambia, evidence of the occurrence of R. felis in domestic dogs, rodents and cat fleas. In particular, the genotype identified from the dogs was identical to a reported genotype detected in cat fleas. Thus, we surmise the possibility of involvement of dogs and cat fleas in the maintenance and transmission of R. felis in Zambia. Besides, the potential of rodents as reservoirs is not negligible even though observed genotypes in Mastomys sp. were slightly diversified from the reported genotype in cat fleas. Interestingly, cat fleas were coinfected with R. asembonensis of unknown pathogenicity with a potential to infect humans. These findings provide evidence of potential circulation of R. felis in Zambia and a risk of human infection. Further studies of potential R. felis infections in humans are needed to understand its epidemiology and ecology.
Abbreviations
- qPCR:
-
quantitative real-time PCR
- Ct:
-
Cycle threshold
- PCR:
-
Polymerase chain reaction
- gltA :
-
Citrate synthetase gene
- OmpA :
-
Outer membrane protein A gene
- OmpB :
-
Outer membrane protein B gene
- bioB :
-
Biotin synthase gene
- RfelB :
-
R. felis unique segment of OmpB
References
Adams JR, Azad AF, Schmidtmann ET. Infection of colonized cat fleas, Ctenocephalides felis (Bouché), with a Rickettsia-like microorganism. Am J Trop Med Hyg. 1990;43:400–9.
Bouyer DH, Stenos J, Crocquet-Valdes P, Moron CG, Popov VL, Zavala-Velazquez JE, et al. Rickettsia felis: molecular characterization of a new member of the spotted fever group. Int J Syst Evol Microbiol. 2001;51:339–47.
Schriefer ME, Sacci JB, Dumler JS, Bullen MG, Azad AF. Identification of a novel rickettsial infection in a patient diagnosed with murine typhus. J Clin Microbiol. 1994;32:949–54.
Fournier PE, Dumler JS, Greub G, Zhang J, Wu Y, Raoult D. Gene sequence-based criteria for identification of new Rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J Clin Microbiol. 2003;41:5456–65.
Parola P. Rickettsia felis: from a rare disease in the USA to a common cause of fever in sub-Saharan Africa. Clin Microbiol Infect. 2011;17:996–1000.
Maina AN, Knobel DL, Jiang J, Halliday J, Feikin DR, Cleaveland S, et al. Rickettsia felis infection in febrile patients, western Kenya, 2007–2010. Emerg Infect Dis. 2012;18:328–31.
Mediannikov O, Socolovschi C, Edouard S, Fenollar F, Mouffok N, Bassene H, et al. Common epidemiology of Rickettsia felis infection and malaria, Africa. Emerg Infect Dis. 2013;19:1775–83.
Reif KE, Macaluso KR. Ecology of Rickettsia felis: a review. J Med Entomol. 2009;46:723–36.
Angelakis E, Mediannikov O, Parola P, Raoult D. Rickettsia felis: the complex journey of an emergent human pathogen. Trends Parasitol. 2016;32:554–64.
Pérez-Osorio CE, Zavala-Velázquez JE, León JJA, Zavala-Castro JE. Rickettsia felis as emergent global threat for humans. Emerg Infect Dis. 2008;14:1019–23.
Renvoisé A, Mediannikov O, Raoult D. Old and new tick-borne rickettsioses. Int Health. 2009;1:17–25.
Rakotonanahary RJL, Harrison A, Maina AN, Jiang J, Richards AL, Rajerison M, et al. Molecular and serological evidence of flea-associated typhus group and spotted fever group rickettsial infections in Madagascar. Parasit Vectors. 2017;10:125.
Abarca K, López J, Acosta-Jamett G, Martínez-Valdebenito C. Rickettsia felis in Rhipicephalus sanguineus from two distant Chilean cities. Vector-Borne Zoonotic Dis. 2013;13:607–9.
Tsui P-Y, Tsai K-H, Weng M-H, Hung Y-W, Liu Y-T, Hu K-Y, et al. Molecular detection and characterization of spotted fever group rickettsiae in Taiwan. Am J Trop Med Hyg. 2007;77:883–90.
Thepparit C, Sunyakumthorn P, Guillotte ML, Popov VL, Foil LD, Macaluso KR. Isolation of a rickettsial pathogen from a non-hematophagous arthropod. PLoS One. 2011;6:e16396.
Socolovschi C, Pagés F, Raoult D. Rickettsia felis in Aedes albopictus mosquitoes, Libreville, Gabon. Emerg Infect Dis. 2012;18:1687–9.
Dieme C, Bechah Y, Socolovschi C, Audoly G, Berenger J-M, Faye O, et al. Transmission potential of Rickettsia felis infection by Anopheles gambiae mosquitoes. Proc Natl Acad Sci USA. 2015;112:8088–93.
Hii SF, Kopp SR, Abdad MY, Thompson MF, O’Leary CA, Rees RL, et al. Molecular evidence supports the role of dogs as potential reservoirs for Rickettsia felis. Vector Borne Zoonotic Dis. 2011;11:1007–12.
Gracia MJ, Marcén JM, Pinal R, Calvete C, Rodes D. Prevalence of Rickettsia and Bartonella species in Spanish cats and their fleas. J Vector Ecol. 2015;40:233–9.
Panti-May JA, Torres-Castro M, Hernández-Betancourt S, Dzul-Rosado K, Zavala-Castro J, López-Avila K, et al. Detection of Rickettsia felis in wild mammals from three municipalities in Yucatan, Mexico. EcoHealth. 2015;12:523–7.
Sashika M, Abe G, Matsumoto K, Inokuma H. Molecular survey of rickettsial agents in feral raccoons (Procyon lotor) in Hokkaido, Japan. Jpn J Infect Dis. 2010;63:353–4.
Boostrom A, Beier MS, Macaluso JA, Macaluso KR, Hayes DS, Radulovic S, et al. Geographic association of Rickettsia felis-infected opossums with human murine typhus, Texas. Emerg Infect Dis. 2002;8:549–54.
Okabayashi T, Hasebe F, Samui KL, Mweene AS, Pandey SG, Yanase T, et al. Short report: prevalence of antibodies against spotted fever, murine typhus, and Q fever rickettsiae in humans living in Zambia. Am J Trop Med Hyg. 1999;61:70–2.
Znazen A, Rolain JM, Hammami N, Hammami A, Jemaa M, Raoult D. Rickettsia felis infection, Tunisia. Emerg Infect Dis. 2006;12:138–40.
Nakayima J, Hayashida K, Nakao R, Ishii A, Ogawa H, Nakamura I, et al. Detection and characterization of zoonotic pathogens of free-ranging non-human primates from Zambia. Parasit Vectors. 2014;7:490.
Masaninga F, Chanda E, Chanda-Kapata P, Hamainza B, Masendu HT, Kamuliwo M, et al. Review of the malaria epidemiology and trends in Zambia. Asian Pac J Trop Biomed. 2013;3:89–94.
Richards AL, Jiang J, Omulo S, Dare R, Abdirahman K, Ali A, et al. Human infection with Rickettsia felis, Kenya. Emerg Infect Dis. 2010;16:1081–6.
Nakamura I, Hangʼombe BM, Sawa H, Kobayashi S, Orba Y, Ishii A, et al. Cross-reactivity of secondary antibodies against African rodents and application for sero-surveillance. J Vet Med Sci. 2013;75:819–25.
Marcos P, Costa J. Ctenocephalides felis felis vs. Ctenocephalides canis (Siphonaptera: Pulicidae): some issues in correctly identify these species. Rev Bras Parasitol Vet. 2012;21:3456.
Wa G, Ohashi N, Aochi M, Ritu W, Wu DX, Yoshikawa Y, et al. Rickettsiae in ticks, Japan, 2007–2011. Emerg Infect Dis. 2013;19:338–40.
Socolovschi C, Mediannikov O, Sokhna C, Tall A, Diatta G, Bassene H, et al. Rickettsia felis-associated uneruptive fever, Senegal. Emerg Infect Dis. 2010;16:1140–2.
Odhiambo AM, Maina AN, Taylor ML, Jiang J, Richards AL. Development and validation of a quantitative real-time polymerase chain reaction assay specific for the detection of Rickettsia felis and not Rickettsia felis-like organisms. Vector Borne Zoonotic Dis. 2014;14:476–81.
Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chapter 2:Unit 2.3.
Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Socolovschi C, Pages F, Ndiath MO, Ratmanov P, Raoult D. Rickettsia species in African Anopheles mosquitoes. PLoS One. 2012;7:e48254.
Oteo JA, Portillo A, Santibáñez S, Blanco JR, Pérez-Martínez L, Ibarra V. Cluster of cases of human Rickettsia felis infection from southern Europe (Spain) diagnosed by PCR. J Clin Microbiol. 2006;44:2669–71.
Rust MK. The biology and ecology of cat fleas and advancements in their pest management: a review. Insects. 2017;8:118.
Hii S-F, Kopp SR, Thompson MF, O’Leary CA, Rees RL, Traub RJ. Molecular evidence of Rickettsia felis infection in dogs from Northern Territory, Australia. Parasit Vectors. 2011;4(198):39.
Silva AB, Vizzoni VF, Costa AP, Costa FB, Moraes-Filho J, Labruna MB, et al. First report of a Rickettsia asembonensis related infecting fleas in Brazil. Acta Trop. 2017;172:44–9.
Oteo JA, Portillo A, Portero F, Zavala-Castro J, Venzal JM, Labruna MB. “Candidatus Rickettsia asemboensis” and Wolbachia spp. in Ctenocephalides felis and Pulex irritans fleas removed from dogs in Ecuador. Parasit Vectors. 2014;7:455.
Sansyzbayev Y, Nurmakhanov T, Berdibekov A, Vilkova A, Yeskhodzhayev O, St. John HK, et al. Survey for rickettsiae within fleas of great gerbils, Almaty Oblast, Kazakhstan. Vector Borne Zoonotic Dis. 2016;17:172–8.
Rzotkiewicz S, Gutiérrez R, Krasnov BR, Morick D, Khokhlova IS, Nachum-Biala Y, et al. Novel evidence suggests that a “Rickettsia felis-like” organism is an endosymbiont of the desert flea, Xenopsylla ramesis. Mol Ecol. 2015;24:1364–73.
Bai Y, Osikowicz LM, Kosoy MY, Eisen RJ, Atiku LA, Mpanga JT, et al. Comparison of zoonotic bacterial agents in fleas collected from small mammals or host-seeking fleas from a Ugandan region where plague is endemic. mSphere. 2017;2:e00402–17.
Jiang J, Maina AN, Knobel DL, Cleaveland S, Laudisoit A, Wamburu K, et al. Molecular detection of Rickettsia felis and Candidatus Rickettsia asemboensis in fleas from human habitats, Asembo, Kenya. Vector Borne Zoonotic Dis. 2013;13:550–8.
Maina AN, Luce-Fedrow A, Omulo S, Hang J, Chan TC, Ade F, et al. Isolation and characterization of a novel Rickettsia species (Rickettsia asembonensis sp. nov.) obtained from cat fleas (Ctenocephalides felis). Int J Syst Evol Microbiol. 2016;66:4512–7.
Han BA, Schmidt JP, Bowden SE, Drake JM. Rodent reservoirs of future zoonotic diseases. Proc Natl Acad Sci USA. 2015;112:7039–44.
Gajda E, Hildebrand J, Sprong H, Buńkowska-Gawlik K, Perec-Matysiak A, Coipan EC. Spotted fever rickettsiae in wild-living rodents from south-western Poland. Parasit Vectors. 2017;10:413.
Minichová L, Hamšíková Z, Mahríková L, Slovák M, Kocianová E, Kazimírová M, et al. Molecular evidence of Rickettsia spp. in ixodid ticks and rodents in suburban, natural and rural habitats in Slovakia. Parasit Vectors. 2017;10:158.
Eremeeva ME, Warashina WR, Sturgeon MM, Buchholz AE, Olmsted GK, Park SY, et al. Rickettsia typhi and R. felis in rat fleas (Xenopsylla cheopis), Oahu, Hawaii. Emerg Infect Dis. 2008;14:1613–5.
Bonwitt J, Sáez AM, Lamin J, Ansumana R, Dawson M, Buanie J, et al. At home with Mastomys and Rattus: human-rodent interactions and potential for primary transmission of Lassa virus in domestic spaces. Am J Trop Med Hyg. 2017;96:935–43.
Acknowledgments
The authors deeply acknowledge Ms. Naoko Kawai for the technical assistance during laboratory experiments and Mr. Alex Kiarie Gaithuma for critical advice.
Funding
This study was funded by AMED programme of the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID; JP18fm0108008) and the International Collaborative Research Programme for Tackling NTD (Neglected Tropical Disease) Challenges in African countries (JP18jm0510001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The data sets supporting the conclusions of this article are included within the article and its additional file. The newly generated DNA sequences were submitted to the DNA Data Bank of Japan (DDBJ) (http://www.ddbj.nig.ac.jp) under the accession numbers LC431490–LC431502.
Authors’ contributions
LCM, KH performed laboratory experiments after DNA extraction. LCM analyzed the data and wrote the manuscript. CS, YE, IN, CK, RN, ASM and ML collected samples, identified sampled species and extracted DNA. CS, KH, JY and BN designed the study, supervised the work and critically revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The sampling of rodents and mosquitoes was conducted under the permission from the Zambia Wildlife Authority (ZAWA), and the use of dog blood was approved by the University of Zambia Biomedical Research Ethics Committee (Ref. No. ZNPW8/27/1 and Ref. No. 016-02-18 respectively).
Consent of publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1: Table S1.
Criteria for Rickettsia species detection and annotation. Table S2. Detailed description of mosquito species sampled and analyzed, and sampling places in Zambia.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Moonga, L.C., Hayashida, K., Nakao, R. et al. Molecular detection of Rickettsia felis in dogs, rodents and cat fleas in Zambia. Parasites Vectors 12, 168 (2019). https://doi.org/10.1186/s13071-019-3435-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-019-3435-6