Abstract
Background
Helicobacter pylori (H. pylori) infection has been recognized as a significant threat for gastric cancer. However, studies that investigated the oncogenic factors and antimicrobial resistance of H. pylori in Egyptian isolates with gastric cancer are rare. The current study aimed to examine: (1) The pattern of antimicrobial resistance of H. pylori isolates of Egyptian gastric cancer patients, and (2) the prevalence of Cytotoxin-associated gene A (CagA).
Methods
Samples were collected from patients with gastric cancer. Isolation of H. pylori was performed using Columbia blood agar supplemented with 10% horse blood, and selective supplement of H. pylori for 3 to 5 days at 37 °C under microaerophilic condition. Isolates were identified by biochemical traits of H. pylori: oxidase, urease and catalase tests. Antimicrobial susceptibility of H. pylori isolates was examined against five antimicrobial agents using disc diffusion method. After that, extraction of DNA and Polymerase Chain Reaction (PCR) were performed to amplify the target genes.
Results
Twelve samples were collected from six males and six females Egyptian patients with cancer with an age range from 22 to 65 years. These cases are scarce and samples were collected over a period of almost eleven months. All isolates were confirmed as positive H. pylori through colony morphology and biochemical tests. The most effective antibiotic found was ciprofloxacin whereas all isolates showed resistance to metronidazole and erythromycin. The target CagA oncogene gene with expected product size was reported and seven (out of twelve) isolates (58%) were identified as CagA positive.
Conclusion
The current study is unique in two main aspects. First, it reported the pattern of antimicrobial susceptibility and prevalence of CagA gene in H. pylori from Egyptian patients. Second, it exclusively recruited isolates from gastric cancer patients which were confirmed by clinical and laparoscopic examination. The moderately high prevalence of CagA gene in Egyptian cancer patients calls for more vigilance against that oncogene.
Similar content being viewed by others
Background
Helicobacter .pylori (H.pylori) is a global issue with increasing rates of infection making it one of the most damaging human pathogens [1, 2]. Infection caused by H. pylori leads to different gastrointestinal disorders including gastritis, gastric ulcer and gastric cancer [3, 4]. However, H. pylori is considered a poor man’s gut pathogen [3] because it has been mainly reported in isolates from developing countries [1, 2, 5]. Recently, World Health Organization (WHO) has classified H. pylori as Class I Carcinogen and a risk factor for gastric cancer, which is acknowledged worldwide as 2nd highest cause of deaths related to cancer [6, 7]. The detection of H. pylori is essential and therefore different types of tests are performed to identify H. pylori in clinical samples. Some tests, that involve an endoscopy to have biopsy samples like urease testing, bacterial culture and PCR [6, 8, 9] have shown excellent sensitivity and specificity for preliminary detection among adults.
One of the key genes of H. pylori is Cytotoxin-associated gene A (Cag A). The translocation of the CagA is encoded by Type IV Secretion System (T4SS) and is associated with gastric cancer [10, 11]. This relationship between H pylori and gastric cancer was interpreted by the injection of CagA protein into epithelial cells through T4SS system, which binds to several cellular proteins and leads to dysregulation of cell division and carcinogenesis [12].
H. pylori is mainly treated through proton pump inhibitors (PPIs) and antimicrobial agents such as amoxicillin, metronidazole or erythromycin. But the ever-increasing resistance against antibiotics reduces the effectiveness of any treatment involving these therapies [13]. H. pylori has a variable antimicrobial sensitivity pattern, depending on the geographical area and the occurrence of H. pylori resistance that reduces the success of first line treatment. Many studies have been reported on H. pylori isolates from different parts of the worlds, including Germany [14], India [15], Brazil [16, 17], Venezuela [18], Chile [19], Colombia [20], Iran [21], Indonesia [22], and Pakistan [23]. In Arabian isolates, limited reports have been published on the antimicrobial resistance and virulence factors of H. pylori in UAE [24] and Kuwait [25, 26]. Though the Egyptian population represents almost one fourth of the Arab nations, few studies have documented the genetic profile and antibiotic sensitivity of H. pylori from Egyptian isolates.
In Egypt, a high prevalence of H. pylori infections has been reported, ranging from 70% in the general population [27], 73% among school children [28], up to 88% in patients with chronic active HCV [29]. With respect to genotypes, CagA positive H. pylori strains in Egyptian isolates was not only associated with gastritis [30], gastric cancer [31], but was reported as a risk factor for ischemic heart diseases. [32]. Studies have also reported contradicting findings with respect to resistance of H. pylori to various antimicrobial protocols in Egyptian hospitals. For instance, resistance to Metronidazole ranged from 25% [33] to 100% [34].
The aim of the present study was two-fold as it examined: (1) The pattern of antimicrobial resistance of H. pylori isolates among gastric cancer patients, and (2) the prevalence of onco-protien CagA gene in isolates by PCR in Egypt.
Methods
Sample collection
This study was conducted in the period from November 2014 to September 2015 at Kasr El-Aini Faculty of Medicine, Cairo University, Egypt. Written consent was obtained from all participants. Gastric biopsy specimens were collected under aseptic conditions and were kept in selective tryptic soy broth as transport media. They were then preserved in the laboratory of microbiology for further processing, as recommended by Siu and colleagues [35].
Patients’ inclusion and exclusion criteria
As per the aim of the study, we recruited only those patients who were diagnosed with gastric cancer and were positive for H. pylori, based on laparoscopic and clinical examination. The patients who had other primary malignancies, or had received proton pump inhibitors or antimicrobial treatment for eradication of H. pylori over the previous three months were excluded from the study.
Isolation of H. pylori
We followed the protocol of Yamaoka and colleagues [36] for the isolation of H. pylori, using Columbia blood agar supplemented with 10% horse blood, and selective supplement of H. pylori (Dent supplement, Oxoid, UK). Then, we incubated the inoculated plates for 3 to 5 days at 37 °C under microaerophilic condition without catalyst using Campylobacter gas kit (Oxoid, UK). According to Oskouei and colleagues [37].
Identification of H. pylori
We followed the protocol of Owen and his colleagues [38] in using the gold standard for identifying the salient cultural characteristics of H. pylori, such as morphology of colonies including shape, texture, margin and size. All slides were further microscopically examined for red curved and straight rods. The H. pylori was identified by its biochemical profile, according to Yamaoka and colleagues [36], such as oxidase, catalase and urease reactions.
Storage of strains
Cultures were stored in a deep freezer at − 80 °C in a sterile Brain Heart Infusion (BHI) (Oxoid - UK), supplemented with 20% glycerol (Sigma Chemical Co. - UK).
Antimicrobial susceptibility pattern
The antimicrobial susceptibility of H. pylori isolates was examined using disc diffusion method against five antimicrobial agents namely; amoxicillin (10 μg), metronidazole (5 μg), tetracycline (30 μg), ciprofloxacin (5 μg) and erythromycin (10 μg). Under microaerophilic condition, the antimicrobial discs were aseptically placed on the dried surface of Muller-Hinton’s agar (MHA) (Oxoid, UK) with 10% horse blood incubated at 37 °C for 72 h. Antimicrobial susceptibility testing to determine zones of inhibition was conformed to the standard of the Clinical and Laboratory Standard Institute (CLSI) with little modification according to previous studies with similar methodology, where a zone size < 25 mm was evaluated as resistant for amoxicillin, > 16 mm for metronidazole resistance [39], > 30 mm for tetracycline, > 17 mm for ciprofloxacin [40] and > 19 mm for erythromycin resistance [41].
DNA extraction and PCR amplification
Extraction of DNA from H. pylori isolates was done from freshly harvested bacterial cells. The DNA was extracted using Qiagen’s QIAamp DNA Mini Kit (Qiagen, Germany) according to manufacturer specifications. Next, all samples were tested by glmM gene to verify H. pylori strains. The screening of CagA was performed by a reaction mixture that contained 1 μL of primer, 1 μL of genomic DNA, 12.5 μL PCR MasterMix, and ddH2O to a total volume of 25 μL. The protocol of PCR was performed starting with 5 min initial denaturation at 95 °C, 30 cycles of 30 s at 94 °C, 30 s at 52 °C, 30 s at 72 °C and a final extension of 72 °C for 5 min. PCR product was detected by gel electrophoresis. The target genes used in the study are listed in Table 1.
Extracted DNA from colonies of H. pylori ATCC 43504 was used as a positive control, while distilled water served as a negative one.
Results
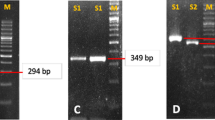
A total of twelve samples (six males and six females) were collected from patients with an age range of 22 to 65 years as per the selection criteria. All isolates showed positive biochemical traits of H. pylori. The twelve isolates were confirmed as H. pylori by amplification of glmM gene using PCR, as demonstrated in Fig. 1.
With respect to antimicrobial sensitivity of H. pylori isolates of Egyptian cancer patients, as listed in Table 2, the most potent antibiotic tested was ciprofloxacin, 10 isolates out of 12 (83% sensitive), the second effective antibiotic was tetracycline, 9 out of 12 (75%), whereas one isolate (8%) was sensitive to amoxicillin. All the isolated strains were resistant to metronidazole and erythromycin.
With respect to the second aim of the study, the prevalence of onco-protien CagA gene in the isolates was confirmed by PCR, as described in the Methodology section. Figure 1 demonstrates that seven (out of twelve) isolates (58%) were identified as CagA positive Fig. 2.
Discussion
The current study aimed to report the antimicrobial sensitivity in H. pylori isolates from Egyptian patients with gastric cancer. Almost all isolates were sensitive to ciprofloxacin and tetracycline and they were resistant to metronidazole and erythromycin. These findings are consistent with those reported by Fathi and colleagues with resistance rates of 100 and 25% for metronidazole and ciprofloxacin respectively in Egypt [34]. In the same vein, amoxicillin and tetracycline were the best options in treating Egyptian patients with H. pylori with an excellent susceptibility of 91 and 82% respectively [42].
In other Middle Eastern countries, resistance to metronidazole in isolates of H. pylori was 78% [43], 57% [44] and 62% [45] in Saudi Arabia, Bahrain and United Arab Emirates (UAE), respectively. While only 2% of isolates were resistant to tetracycline in Saudi Arabia [43], almost no resistance was reported to tetracycline in Bahrain or UAE [44, 45]. A systemic review from Iran reported a growing antimicrobial resistance of H. pylori, particularly against metronidazole (62%), amoxicillin (16%), erythromycin (15%) and tetracycline (12%) [46, 47]. Rasheed and his colleagues reported a similar antibiotic sensitivity profile in H. pylori isolates from Pakistani patients, with a relatively low resistance percentages of ciprofloxacin and tetracycline, at 13 and 4% respectively and a high resistance to metronidazole and clarithromycin, at 74 and 48% correspondingly. The alarming sign was that almost 93% of Pakistani isolates showed resistance to one or more antibiotics.
As evident in literature that the H. pylori antibiotic resistance against metronidazole and clarithromycin is principally challenging, other treatment approaches have been suggested, which are still under investigation. These include complementary probiotic therapy with Lactobacillus that could be feasible alternate eradication therapy [48].
The second aim of the study is related to explore the prevalence of CagA gene in isolates by PCR in Egypt. The notorious reputation of CagA gene as an oncogenic protein was echoed by the results of the study, where almost 60% of isolates from gastric cancer patients were positive CagA. This is relatively higher than the prevalence of CagA, as reported by other studies, such as 46% [49], and 50% [50] in Egyptian isolates of H. pylori. While in other countries in the Middle East, CagA gene was even lower in H. pylori isolates from Kuwait (41%) [51] and Jordan (26%) [52]. The moderately high prevalence of CagA gene in Egyptian patients with gastric cancer calls for more vigilance against this oncogene.
The correlation of CagA gene with cancer was established in H. pylori isolates from Turkish patients [53]. However, in South Mexico, a study reported no association between CagA genotype and gastric cancer patients [54]. The reports of different studies across various geographical regions reinforce the unpredictability of the expression of CagA gene of H Pylori across different populations. For instance, the CagA gene showed strong signatures in isolates from Venezuelan (Amer-Indian) populations, but not in Asian ones [12] The variability of adaptation models of CagA gene in different ethnic groups shows the effect of host genetics or other lifestyle patterns that moderate its expression. Generally speaking, peptic ulcer diseases were remarkably higher in patients with CagA positive H pylori strains than the ones with CagA negative strains, but the presence of CagA gene has not been associated with severity [26]. Still there is a need to explore further the moderating correlation and presence of CagA gene in gastric cancer patients.
Salient features of the study
The current study is unique in two main aspects. First, it reported the pattern of antimicrobial susceptibility and prevalence of CagA gene in H. pylori among Egyptian patients. Second, it exclusively recruited gastric cancer patients who had been confirmed by clinical and laparoscopic examination. As these cases are scarce, they were collected over a period of almost eleven months.
Venues for further research
There’s a need to perform DNA sequencing of the same fragment obtained from each isolate with relevance to their antibiotic resistance pattern to confirm the results and explore the mechanism of antibiotic resistance in Egyptian isolates of H. pylori. A comparison can then be reported between isolates from different geographical locations, particularly in the Middle Eastern region. Moreover, regarding the correlation of CagA gene with gastric cancer, further research is needed to explore its mechanism and variables that moderate its effect in the host. This approach might facilitate to gain insight into the profile of antibiotic resistance pattern and CagA gene in Egyptian isolates of H. pylori.
Conclusion
The current study reported high susceptibility of H. pylori to ciprofloxacin and tetracycline, which is promising to eradicate the infection in Egypt. The pattern of antimicrobial susceptibility among gastric cancer patients has to be frequently investigated to guide clinicians to choose effective antibiotics for H. pylori infections and monitor the antibiotics policy. The results show that CagA gene was present in almost 60% of H. pylori isolates from cancer patients. Therefore, it is necessary to screen it in all cases with H. pylori infection as its association with threating prognosis calls for more vigilance against this oncogene.
Abbreviations
- BHI:
-
Brain Heart Infusion
- CagA:
-
Cytotoxin-associated gene A
- CLSI:
-
Clinical & Laboratory Standards Institute
- MHA:
-
Muller-Hinton’s agar
- PCR:
-
Polymerase Chain Reaction
- T4SS:
-
Type IV Secretion System
- WHO:
-
World Health Organization
References
Eusebi LH, Zagari RM, Bazzoli F. Epidemiology of Helicobacter pylori Infection. Helicobacter. 2014:1–5.
Nagy P, Johansson S. Molloy-bland M. Gut Pathog: Systematic review of time trends in the prevalence of Helicobacter pylori infection in China and the USA; 2016.
Khalifa MM, Sharaf RR, Aziz RK. Helicobacter pylori: a poor man’s gut pathogen? Gut Pathog. 2010.
Ahmed N, Tenguria S, Nandanwar N. Helicobacter pylori--a seasoned pathogen by any other name. Gut Pathog. 2009;1:24.
Hunt RH, Xiao SD, Megraud F, Leon-Barua R, Bazzoli F, van der Merwe S, et al. Helicobacter pylori in developing countries. World gastroenterology organisation global guideline. J. Gastrointest. Liver Dis. 2011:299–304.
Wang F, Meng W, Wang B, Qiao L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 2014:196–202.
Wroblewski LE, Peek RM, Wilson KT. Helicobacter pylori and gastric cancer: Factors that modulate disease risk. Clin. Microbiol. Rev. 2010:713–39.
Kienesberger S, Perez-Perez GI, Rivera-Correa JL, et al. Gut Pathog. 2012;4:9. https://doi.org/10.1186/1757-4749-4-9.
Kusters JG, Van Vliet AHM, Kuipers EJ. Pathogenesis of helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449–90.
Busler VJ, Torres VJ, McClain MS, Tirado O, Friedman DB, Cover TL. Protein-protein interactions among helicobacter pylori cag proteins. J Bacteriol. 2006;188:4787–800.
Vannini A, Roncarati D, Spinsanti M, Scarlato V, Danielli A. In depth analysis of the Helicobacter pylori cag pathogenicity island transcriptional responses. PLoS One [Internet]. 2014;9:e98416. Available from: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0098416
Delgado-Rosado G, Dominguez-Bello M, Massey SE. Positive selection on a bacterial oncoprotein associated with gastric cancer. Gut Pathog. [Internet]. BioMed Central; 2011 [cited 2018 Jan 19];3:18. Available from: http://gutpathogens.biomedcentral.com/articles/10.1186/1757-4749-3-18
Safavi M, Sabourian R, Foroumadi A. Treatment of helicobacter pylori infection: current and future insights. World J Clin Cases. 2016;4:5–19.
Michel A, Pawlita M, Boeing H, Gissmann L, Waterboer T. Helicobacter pylori antibody patterns in Germany: a cross-sectional population study. Gut Pathog. 2014;6:10. https://doi.org/10.1186/1757-4749-6-10.
De R, Kundu P, Swarnakar S, Ramamurthy T, Chowdhury A, Nair GB, et al. Antimicrobial activity of curcumin against helicobacter pylori isolates from India and during infections in mice. Antimicrob Agents Chemother. 2009;53:1592–7.
Oliveira JG de, Ferreira CHT, Camerin ACS, Rota CA, Meurer L, Silveira TR da. Prevalence of infection with cagA-positive Helicobacter pylori strains among children and adolescents in southern Brazil. Arq. Gastroenterol. [Internet]. [cited 2018 Jan 14];51:180–185. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25296076.
Bonacorsi C, da Fonseca LM, Raddi MSG, Kitagawa RR, Vilegas W. Comparison of Brazilian Plants Used to Treat Gastritis on the Oxidative Burst of Helicobacter pylori-Stimulated Neutrophil. Evid. Based. Complement. Alternat. Med. [Internet]. 2013 [cited 2018 Jan 14];2013:851621. Available from: http://www.hindawi.com/journals/ecam/2013/851621/
Contreras M, Abrante L, Salazar V, Reyes N, García-Amado MA, Fernández-Delgado M, et al. Heterogeneity of cag genotypes of helicobacter pylori in the esophageal mucosa of dyspeptic patients and its relation to histopathological outcomes. Int J Infect Dis. 2014;26:91–5.
Toledo H, López-Solís R. Tetracycline resistance in Chilean clinical isolates of helicobacter pylori. J Antimicrob Chemother. 2009;65:470–3.
Gutiérrez-Escobar AJ, Trujillo E, Acevedo O, Bravo MM. Phylogenomics of Colombian helicobacter pylori isolates. Gut Pathog. 2017;9:52. https://doi.org/10.1186/s13099-017-0201-1.
Talebi Bezmin Abadi A, Mohabbati Mobarez A. High Prevalence of Helicobacter pylori hopQ II Genotype Isolated from Iranian Patients with Gastroduodenal Disorders. J. Pathog. [Internet]. 2014;2014:842469; https://doi.org/10.1155/2014/842469.
Miftahussurur M, Tuda J, Suzuki R, Kido Y, Kawamoto F, Matsuda M, et al. Extremely low helicobacter pylori prevalence in North Sulawesi. Indonesia and identification of a Maori-tribe type strain: A cross sectional study Gut Pathog. 2014;6:42. https://doi.org/10.1186/s13099-014-0042-0.
Rasheed F, Campbell BJ, Alfizah H, Varro A, Zahra R, Yamaoka Y, et al. Analysis of clinical isolates of helicobacter pylori in Pakistan reveals high degrees of pathogenicity and high frequencies of antibiotic resistance. Helicobacter. 2014;19:387–99.
Alfaresi MS, Elkoush AA. Characterization of clarithromycin resistance in isolates of helicobacter pylori from the UAE. Indian J Gastroenterol. 2010;29:116–20.
Alazmi WM, Siddique I, Alateeqi N, Al-Nakib B. Prevalence of Helicobacter pylori infection among new outpatients with dyspepsia in Kuwait. BMC Gastroenterol. [Internet]. 2010;10. Available from: https://bmcgastroenterol.biomedcentral.com/articles/10.1186/1471-230X-10-14.
Siddique I, Al-Qabandi A, Al-Ali J, Alazmi W, Memon A, Mustafa AS, et al. Association between helicobacter pylori genotypes and severity of chronic gastritis, peptic ulcer disease and gastric mucosal interleukin-8 levels: evidence from a study in the Middle East. Gut Pathog. 2014;6:41. https://doi.org/10.1186/s13099-014-0041-1.
Ghaith D, Elzahry M, Mostafa G, Mostafa S, Elsherif R, Ramzy I. Mutations affecting domain V of the 23S rRNA gene in Helicobacter pylori from Cairo, Egypt. J. Chemother. [Internet]. 2016 [cited 2018 Jun 14];28:367–370. Available from: https://www.tandfonline.com/doi/full/10.1179/1973947815Y.0000000067
Mohammad MA, Hussein L, Coward A, Jackson SJ. Prevalence of helicobacter pylori infection among Egyptian children: impact of social background and effect on growth. Public Health Nutr. 2008;11:230–6.
Hanafy AS, El Hawary AT, Hamed EF, Hassaneen AM. Impact of Helicobacter pylori eradication on refractory thrombocytopenia in patients with chronic HCV awaiting antiviral therapy. Eur. J. Clin. Microbiol. Infect. Dis. [Internet]. Springer Berlin Heidelberg; 2016 [cited 2018 Jun 14];35:1171–6. Available from: http://link.springer.com/10.1007/s10096-016-2650-8
El-Fakhry AA, El-Daker MA, Badr RI, El-Nady GM, Mesbah MR, Youssef T, et al. Association of the CagA gene positive Helicobacter pylori and tissue levels of interleukin-17 and interleukin-8 in gastric ulcer patients. Egypt. J. Immunol. [Internet]. 2012 [cited 2018 Jun 14];19:51–62. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23888551.
Said Essa A, Alaa Eldeen Nouh M, Mohammed Ghaniam N, Graham DY, Said Sabry H. Prevalence of cagA in relation to clinical presentation of Helicobacter pylori infection in Egypt. Scand. J. Infect. Dis. [Internet]. 2008 [cited 2018 Jun 14];40:730–733. Available from: http://www.tandfonline.com/doi/full/10.1080/00365540802023725
El-Mashad N, El-Emshaty WM, Arfat MS, Koura BA, Metwally SS. Relation of Cag-A-positive Helicobacter pylori strain and some inflammatory markers in patients with ischemic heart diseases. Egypt. J. Immunol. [Internet]. 2009 [cited 2018 Jun 14];16:39–47. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20726321.
Diab M, El-Shenawy A, El-Ghannam M, Salem D, Abdelnasser M, Shaheen M, et al. Detection of antimicrobial resistance genes of helicobacter pylori strains to clarithromycin, metronidazole, amoxicillin and tetracycline among Egyptian patients. Egypt. J. Med. Hum. Genet. 2018;
Fathi MS, RF EL-f, Hassan RA, El-Arab ME. Genotypic and phenotypic patterns of antimicrobial susceptibility of Helicobacter pylori strains among Egyptian patients. Egypt. J. Med. Hum. Genet. 2013;
Siu LK, Leung WK, Cheng AF, Sung JY, Ling TK, Ling JM, et al. Evaluation of a selective transport medium for gastric biopsy specimens to be cultured for Helicobacter pylori. J. Clin. Microbiol. [Internet]. 1998 [cited 2018 Jan 14];36:3048–3050. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9738066.
Yamaoka Y, Kodama T, Gutierrez O, Kim JG, Kashima K, Graham DY. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J. Clin. Microbiol. [Internet]. 1999 [cited 2018 Jan 14];37:2274–2279. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10364597.
Oskouei DD, Bekmen N, Ellidokuz H, Yılmaz O. Evaluation of different cryoprotective agents in maintenance of viability of Helicobacter pylori in stock culture media. Braz. J. Microbiol. [Internet]. 2010 [cited 2018 Jan 14];41:1038–1046. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24031584.
Owen RJ, Slater ER, Xerry J, Peters TM, Teare EL, Grant A. Development of a scheme for genotyping Helicobacter pylori based on allelic variation in urease subunit genes. J. Clin. Microbiol. [Internet]. 1998 [cited 2018 Jan 14];36:3710–3712. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9817904.
Ogata SK, Gales AC, Kawakami E. Antimicrobial susceptibility testing for Helicobacter pylori isolates from Brazilian children and adolescents: comparing agar dilution, E-test, and disk diffusion. Braz. J. Microbiol. [Internet]. 2014 [cited 2018 Jan 14];45:1439–1448. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25763052.
Ozbey G, Bahcecioglu IH, Acik MN. Prevalence of helicobacter pylori in children in eastern Turkey and molecular typing of isolates. Braz J Microbiol. 2015;46:505–11.
Tanih NF, Okeleye BI, Naidoo N, Clarke AM, Mkwetshana N, Green E, et al. Marked susceptibility of South African Helicobacter pylori strains to ciprofloxacin and amoxicillin: Clinical implications. S Afr Med J. 2010 [cited 2010 Jan 1]; 100:49–52. Available from: https://www.ncbi.nlm.nih.gov/pubmed/20429489.
Hafez R, Hasanein W, Elsayd H, Basha O, Elsayd MA. Antibiotic Resistance and Failure of Eradication of Helicobacter Pylori in Egypt. Egypt. J. Med. Microbiol. 2011;20(2):149–54.
Al-Qurashi AR, El-Morsy F, Al-Quorain AA. Evolution of metronidazole and tetracycline susceptibility pattern in Helicobacter pylori at a hospital in Saudi Arabia. Int. J. Antimicrob. Agents [Internet]. 2001 [cited 2018 Jun 19];17:233–236. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11282271.
Bindayna KM. Antibiotic susceptibilities of Helicobacter pylori. Saudi Med. J. [Internet]. 2001 [cited 2018 Jun 19];22:53–57. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11255612.
Khedmat H, Karbasi-Afshar R, Agah S, Taheri S. Helicobacter pylori Infection in the general population: A Middle Eastern perspective. Casp. J. Intern. Med. [Internet]. Babol University of Medical Sciences; 2013 [cited 2018 Jun 19];4:745–753. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24294467.
Khademi F, Poursina F, Hosseini E, Akbari M, Safaei HG. Helicobacter pylori in Iran: a systematic review on the antibiotic resistance [internet]. Iran. J. Basic med. Sci. Mashhad University of Medical Sciences; 2015 [cited 2018 Jun 19]. p. 2–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25810869.
Yousefi-Avarvand A, Vaez H, Tafaghodi M, Sahebkar AH, Arzanlou M, Khademi F. Antibiotic Resistance of Helicobacter pylori in Iranian Children: A Systematic Review and Meta-Analysis. Microb. Drug Resist. [Internet]. Mary Ann Liebert, Inc. 140 Huguenot Street, 3rd Floor New Rochelle, NY 10801 USA; 2017 [cited 2018 Jun 19];mdr.2017.0292. Available from: http://www.liebertpub.com/doi/10.1089/mdr.2017.0292
Kim SY, Choi DJ, Chung J-W. Antibiotic treatment for helicobacter pylori: is the end coming? World J Gastrointest Pharmacol Ther [Internet]. 2015;6:183–98. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4635158&tool=pmcentrez&rendertype=abstract
El-Khlousy M, Rahman EA, Mostafa S, Bassam A, Elgawad HA, Elnasr MS, et al. Study of the clinical relevance of Helicobacter pylori virulence genes to gastric diseases among Egyptian patients. Arab J. Gastroenterol. [Internet]. 2016 [cited 2018 Jun 14];17:90–94. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1687197916300259
Abu-Taleb AMF, Abdelattef RS, Abdel-Hady AA, Omran FH, El-korashi LA, Abdel-aziz El-hady H, et al. Prevalence of Helicobacter pylori cagA and iceA genes and their association with gastrointestinal diseases. Int J Microbiol [Internet] 2018;2018:1–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29849647%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5907521%0Ahttps://www.hindawi.com/journals/ijmicro/2018/4809093/
Al QA, Mustafa AS, Siddique I, Khajah AK, Madda JP. Junaid TA Distribution of vacA and cagA genotypes of Helicobacter pylori in Kuwait. Acta Trop. 2005;93:283–8.
Nimri LF, Matalka I, Hani KB, Ibrahim M. Helicobacter pylori genotypes identified in gastric biopsy specimens from Jordanian patients. BMC Gastroenterol. 2006;6:27.
Salih BA, Bolek B, Yildiz M, Arikan S. Phylogenetic analysis of Helicobacter pylori cagA gene of Turkish isolates and the association with gastric pathologyGut Pathog. [Internet]. BioMed Central; 2013 [cited 2018 Jan 20];5:33. Available from: http://gutpathogens.biomedcentral.com/articles/10.1186/1757-4749-5-33
Román-Román A, Martínez-Carrillo DN, Atrisco-Morales J, Azúcar-Heziquio JC, Cuevas-Caballero AS, Castañón-Sánchez CA, et al. Helicobacter pylori vacA s1m1 genotype but not cagA or babA2 increase the risk of ulcer and gastric cancer in patients from Southern Mexico. Gut Pathog. [Internet]. BioMed Central; 2017 [cited 2018 Jan 21];9:18. Available from: http://gutpathogens.biomedcentral.com/articles/10.1186/s13099-017-0167-z
Espinoza MGC, Vazquez RG, Mendez IM, Vargas CR, Cerezo SG. Detection of the glmM gene in Helicobacter pylori isolates with a novel primer by PCR. J. Clin. Microbiol. [Internet]. American Society for Microbiology (ASM); 2011 [cited 2018 Jan 21];49:1650–2. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21289140.
Izadi F, Ahmadi A, Ghourchian S, Daneshi A, Memari F, Khadivi E, et al. Detection of helicobacter pylori in benign laryngeal lesions by polymerase chain reaction: a cross sectional study. Infect. Agent. Cancer [Internet]. BioMed Central; 2012 [cited 2018 Jan 21];7:10. Available from: http://infectagentscancer.biomedcentral.com/articles/10.1186/1750-9378-7-10
Acknowledgements
The authors would like to thank Dr. Mubina Rauf for English language editing of the manuscript.
Availability of data and materials
Data and materials have been provided in the main manuscript, where necessary additional information of the study can be made available from the corresponding author on request.
Author information
Authors and Affiliations
Contributions
DME, OMH, MAR conceived of the study, YMR participated in the design of this study; ZAK and EAS coordinate the collection of specimens; EAS and MAR carried out bacterial isolation and identification; DME, MAR, YMR performed the antimicrobial susceptibility; DME and OMH implemented DNA extraction and PCR amplification. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The protocol of this study was approved by the review board of ethics committee of College of Pharmacy of Cairo University and written consents were obtained from all participants.
Consent for publication
“Not applicable”.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Al-Eraky, D.M., Helmy, O.M., Ragab, Y.M. et al. Prevalence of CagA and antimicrobial sensitivity of H. pylori isolates of patients with gastric cancer in Egypt. Infect Agents Cancer 13, 24 (2018). https://doi.org/10.1186/s13027-018-0198-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13027-018-0198-1