Abstract
Background
Genetic risk factors for chemotherapy-induced peripheral neuropathy (CIPN), a major dose-limiting side-effect of paclitaxel, are not well understood.
Methods
We performed a genome-wide association study (GWAS) in 183 paclitaxel-treated patients to identify genetic loci associated with CIPN assessed via comprehensive neuropathy phenotyping tools (patient-reported, clinical and neurological grading scales). Bioinformatic analyses including pathway enrichment and polygenic risk score analysis were used to identify mechanistic pathways of interest.
Results
In total, 77% of the cohort were classified with CIPN (n = 139), with moderate/severe neuropathy in 36%. GWAS was undertaken separately for the three measures of CIPN. GWAS of patient-reported CIPN identified 4 chromosomal regions that exceeded genome-wide significance (rs9846958, chromosome 3; rs117158921, chromosome 18; rs4560447, chromosome 4; rs200091415, chromosome 10). rs4560447 is located within a protein-coding gene, LIMCH1, associated with actin and neural development and expressed in the dorsal root ganglia (DRG). There were additional risk loci that exceeded the statistical threshold for suggestive genome-wide association (P < 1 × 10–5) for all measures. A polygenic risk score calculated from the top 46 ranked SNPs was highly correlated with patient-reported CIPN (r2 = 0.53; P = 1.54 × 10–35). Overlap analysis was performed to identify 3338 genes which were in common between the patient-reported CIPN, neurological grading scale and clinical grading scale GWAS. The common gene set was subsequently analysed for enrichment of gene ontology (GO) and Reactome pathways, identifying a number of pathways, including the axon development pathway (GO:0061564; P = 1.78 × 10–6) and neuronal system (R-HSA-112316; adjusted P = 3.33 × 10–7).
Conclusions
Our findings highlight the potential role of axon development and regeneration pathways in paclitaxel-induced CIPN.
Similar content being viewed by others
Background
Paclitaxel is a highly active chemotherapeutic agent used widely in the treatment of solid tumours [1]. However, chemotherapy-induced peripheral neurotoxicity (CIPN) is a major dose-limiting neurological side-effect of paclitaxel treatment that can persist long-term [2]. CIPN produces sensory and functional abnormalities leading to difficulties with fine motor and balance tasks, increased falls risk, and reduced quality of life [3]. Further, CIPN is a common cause of dose reduction and premature discontinuation, potentially affecting survival outcomes [4]. There are currently no neuroprotective measures to prevent the development of CIPN and no effective treatment options [5]. Importantly, understanding the mechanisms underlying CIPN and identifying which patients are most at-risk are critical to preventing long-term sequelae of treatment with paclitaxel.
Mechanistically, paclitaxel targets microtubules, inhibiting the dynamic assembly and disassembly of β-tubulin, leading to their stabilisation, cell-cycle arrest, and cell death [1]. While this mechanism has been proposed to produce neurotoxicity via disruption to axonal transport [6], growing evidence suggests a range of additional mechanisms, including disruption of neuronal cell metabolism, mitochondrial dysfunction, oxidative stress and neuroinflammation as underlying the development of CIPN [7]. Better understanding of underlying mechanisms of CIPN will be critical to the development of successful preventative and treatment strategies.
There have been substantial efforts to identify genetic profiles associated with heightened CIPN risk, with a range of single nucleotide polymorphisms (SNPs) in genes associated with neural development and structure, drug metabolism and neural repair associated with paclitaxel-induced CIPN [8]. However, there has been limited replication between studies, and there remains a lack of validated genetic associations with paclitaxel-induced CIPN. A key limitation is the lack of consensus regarding appropriate CIPN outcome measures [9]. It has been well documented that patients report greater neuropathy severity than clinicians [10] and that patient-reported outcomes and clinician-reported outcomes provide complementary but different information about CIPN [11]. However, despite this, there has been limited studies incorporating multimodal CIPN outcome measures with comprehensive phenotyping and patient-reported outcome measures.
Ultimately, identification and validation of genetic pathways involved in CIPN will enable characterisation of patients at-risk of significant, persistent toxicity. CIPN risk likely incorporates multiple genes [7] and polygenic models will be required to explain variability in CIPN outcomes rather than reliance on single SNPs. However, such models need to be developed in appropriately phenotyped cohorts. In the present study, we utilised comprehensive CIPN assessment and phenotyping using multiple assessment tools combined with genome-wide association studies (GWAS) and pathway analysis to provide an improved understanding of the genetic variants contributing to paclitaxel-induced CIPN.
Methods
We performed a GWAS on 183 paclitaxel-treated patients with comprehensive neuropathy phenotyping. Bioinformatic analyses, including pathway enrichment and polygenic risk score analysis, were used to identify mechanistic pathways of interest.
Participants and neuropathy assessment
Germline DNA samples, clinical details and detailed neuropathy phenotyping were collected from paclitaxel-treated patients enrolled in observational CIPN cohort studies at Australian cancer centres. Patients assessed for neuropathy status following completion of paclitaxel-based treatment were eligible. Data relating to cancer diagnosis and treatment were recorded from medical records. Ethical approval was granted by the Sydney Local Health District and South-Eastern Sydney Local Health District Human Research Ethics Committees. All patients provided written informed consent to participate.
Patients underwent a clinical and functional CIPN assessment following the completion of paclitaxel chemotherapy. Multiple methods were used to quantify CIPN, including the clinical grading scale National Cancer Institute Common Terminology Criteria for Adverse Events (NCI-CTCAE) version 4.0 sensory neuropathy subscale, which graded CIPN severity as Grade-0 ‘no symptoms’, 1 ‘asymptomatic, not interfering with daily function’, 2 ‘moderate symptoms, limiting daily function’, 3 ‘severe symptoms, limiting daily function and self-care’, and 4 ‘disabling’. The neurological grading scale Total Neuropathy Score–clinical version (TNSc © Johns Hopkins University)[12] was utilized and incorporated six domains (sensory symptoms, motor symptoms, upper and lower limb pinprick and vibration sensibility, lower limb strength, deep tendon reflexes) graded from 0 to 4 (most severe presentation), for a maximum score of 24.
The patient-reported outcome measure European Organization for Research and Treatment of Cancer Quality of Life Questionnaire-Chemotherapy-Induced Peripheral Neuropathy (EORTC QLQ-CIPN20) [13] was utilized and included 20 symptom questions, each rated from “not at all” (1) to “very much” (4) before summation and linear transformation to a 0–100 scale, with higher numbers representing greater CIPN.
Genotyping and quality control
DNA was extracted from blood and genotyped using the Illumina INFINIUM Microarray on GlobalScreeningAssay-24, with coverage of ~ 654,027 fixed markers. Quality control procedures and GWAS were implemented in the free, open‐source whole‐genome association analysis toolset PLINK version1.9 and R statistical software. Following sample quality control, individuals with a heterozygosity rate > 0.03% or sex discrepancy were excluded. SNPs with poor genotype clustering performance, excess missingness > 1%, minor allele frequency < 1%, and out of Hardy Weinberg equilibrium proportions < 1e−6 were removed, leaving 289,351 SNPs for subsequent analysis. The multidimensional scaling approach was used for the correction of population stratification using the 1000 Genomes Project data.
Bioinformatic and statistical analyses
Clinical correlation analysis was undertaken in GraphPad Prism version 9.3.1 for Windows (GraphPad Software, California USA), using Mann–Whitney U tests or Spearman’s correlation coefficients. Normality was assessed using the Shapiro–Wilk test and results were presented as mean and standard deviation or median (interquartile range IQR) or for normally distributed and non-normally distributed data, respectively.
Genome-wide association studies
Within PLINK, linear regression models were fit to predict the association between SNPs and CIPN using continuous variables related to phenotypes of interest. Correlation analyses indicated that age and body mass index (BMI) were significantly associated with all measures of CIPN and were included as co-variates in the subsequent GWASs. Quantile–quantile (Q-Q) plots of the marginal asymptotic P values were evaluated for the remaining population stratification. Sanity checks of individual variants were conducted in SPSS software package v.26.0 to confirm results. SNPs with nominal P values of ≤ 5 × 10–8 were considered to exceed genome-side significance, and P values of < 1 × 10–5 were considered suggestive for genome-wide association. LocusZoom was used to generate Manhattan plots and higher resolution plots of top associated SNPs [14].
PRS analysis
The P values and effect sizes (Beta values) from the top-ranked SNPs identified from the EORTC-QLQ-CIPN20 GWAS were used to calculate the polygenic risk score (PRS) using the PRSice package version 2.3.2 [15]. PRS were computed for a range of P value thresholds from the GWAS top-ranked SNPs. A correlation (r2) P value was calculated between an individual’s PRS and their patient-reported CIPN for each defined P-value threshold in order to identify the optimal panel of SNPs based on the association between the PRS and phenotype. The PRSice algorithm also adjusted for linkage disequilibrium between the SNPs using the Clump function to identify index SNPs used to calculate the PRS.
Candidate variant analysis
We included variants that were reported in previous GWAS studies to be significantly associated with paclitaxel-induced neuropathy for replication using our dataset. LDlinkR [16] computational software was interrogated to identify the optimal proxy variant (r2 < 0.1) with our dataset and to determine whether a SNP of interest lies in a potential regulatory genomic region. LDlinkR, which contains data from the 1000 Genomes Project, searches for proxy and putatively functional variants by exploring linkage disequilibrium (LD) structure in a native R environment.
Pathway enrichment analysis
The PASCAL (Pathway scoring algorithm) [17] was used to provide insight into the biological processes in terms of gene-based P values by aggregating the association signal from GWAS analysis while correcting for LD structure using 1000 Genomes Project data [18]. Functional enrichment (Over-Representation Analysis (ORA)) of Geneontology (Biological Process) and Pathways (Reactome) in our list of genes commonly identified by all three measures of neuropathy was performed using WebGestalt (WEB-based Gene SeT AnaLysis Toolkit).
Results
Clinical characteristics
A total of 183 paclitaxel-treated patients were included in the GWAS analyses (Table 1). The median age was 59 years (range 27–85 years). The majority were breast cancer (n = 103, 56%) or ovarian cancer (n = 41, 23%) patients who completed their paclitaxel-based chemotherapy (median cumulative dose 960 (IQR 240) mg/m2) at a median of 6 months prior to assessment (range 0–59 months). 78 patients received concurrent carboplatin (n = 77) or cisplatin (n = 1).
In total, 77% of the cohort were classified with CIPN (n = 139), with moderate/severe CIPN in 36% (NCI-CTCAE grade 2 + ; n = 65; Additional file 1: Fig. S1). All three measures of CIPN were correlated with each other (NCI-CTCAE and TNSc r = 0.60; NCI and EORTC-QLQ-CIPN20 r = 0.77; TNSc and EORTC-QLQ-CIPN20 r = 0.62; all P < 0.0001). There was no significant difference in CIPN severity by platinum-treatment status (NCI-CTCAE P = 0.628; TNSc P = 0.799; or EORTC-QLQ-CIPN20 score P = 0.598) or time since treatment (≥ 6 vs < 6 months post completion of paclitaxel; NCI-CTCAE P = 0.538; TNSc P = 0.98; EORTC-QLQ-CIPN20 P = 0.726). All three measures of CIPN were significantly associated with age (NCI-CTCAE r = 0.400; TNSc r = 0.401; EORTC-QLQ-CIPN20 r = 0.395; all P < 0.0001) and BMI (NCI-CTCAE r = 0.216; TNSc r = 0.217; EORTC-QLQ-CIPN20 r = 0.283; all P < 0.001).
GWAS identifies four chromosomal regions significantly associated with patient-reported CIPN
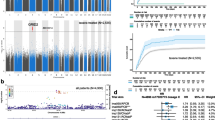
GWAS was undertaken separately for the three measures of CIPN. Notably, the GWAS of patient-reported CIPN (EORTC-QLQ-CIPN20) identified 4 chromosomal regions that exceeded genome-wide significance (Fig. 1A and Table 2). In addition, there were risk loci that exceeded the statistical threshold for suggestive genome-wide association (P < 1 × 10–5) for patient-reported CIPN as well as for both clinical CIPN (NCI-CTCAE) and neurologically graded CIPN (TNSc) (Table 2). Q-Q plots of the expected and observed P values from the GWAS of the three measures of CIPN were generated (Additional file 1: Fig. S2). The 50th percentile genomic control lambda values were ~ 1, indicating that there were no underlying population stratifications in the patient cohort. The functional consequence of the top associated SNPs (P < 10–5) for the patient-reported CIPN GWAS, annotated using Ensembl Variant Effect Predictor (VEP) platform [19], indicated that the majority of SNPs (69%) mapped to known genes, of which 14/37 (38%) were within non-coding RNAs (Fig. 1B, Additional file 2: Table S1). Of note, 18 of these SNPs were considered intergenic (10 kb distal to known genes). Four of the intergenic variants were located within known regulatory elements known as enhancers (rs7536740, rs111669817, rs117378411 and rs728169) Additional file 2: Table S1), while another two intergenic SNPs (rs75263049 and rs77573336) had unknown functions, but had Combined Annotation Dependent Depletion (CADD) scores > 10..
A) Manhattan plot showing the unadjusted P-value for the association of all SNPs with patient-reported CIPN (EORTC-QLQ-CIPN20). GWAS identified four loci that exceeded genome-wide significance of P < 5 × 10–8 (red circles). B) Distribution of top associated SNPs from EORTC-QLQ-CIPN20 GWAS (P < 10–5 cut-off) into functional categories including known genes, non-coding RNAs and intergenic variants within regulatory elements. C) Bar plot for PRS calculated for each P-value range and their correlation with the patient-reported scores. The corresponding number of SNPs that fall within each threshold is indicated in brackets
Fine mapping of patient-reported CIPN GWAS loci and eQTL co-localisation
A high-resolution view of the genomic landscape of the four SNPs from the GWAS of patient-reported CIPN (EORTC-QLQ-CIPN20) that achieved genome-wide significance and the closest known genes was undertaken via LocusZoom (Additional file 1: Fig. S3). Potential expression quantitative trait loci (eQTL) within each top associated chromosomal region were identified on the basis of available gene expression data for the tibial nerve tissue as in [20] and RNA-Seq data for the DRG [21] (Table 2).
As shown in Additional file 1: Fig. S3A, rs4560447 on chromosome 4 is located within a protein-coding gene, LIMCH1, which encodes the LIM and calponin homology domains-containing protein associated with actin stress fibres [22]. Interrogation of Genotype-Tissue Expression (GTEx) database [23] using the LDexpress Tool [24], confirmed that a proxy variant for rs4560447 (rs79278739 with D’ = 1.0 and r2 = 0.817) is a significant eQTL for LIMCH1 in coronary artery tissue, but no eQTL were detected for the tibial nerve (Table 3). However, LIMCH1 is expressed in the DRG with the FPKM (Fragments Per Kilobase of transcript per Million mapped reads) value of 16.9.
Another 2 top associated variants, rs117158921 on chromosome 18 and rs200091415 on chromosome 10, mapped to two long non-coding RNAs (LncRNA) AC124254.2 and FAM238B, respectively (Additional file 1: Fig. S3B and C, Table 3). The function of these two LncRNAs are currently unknown. There were no proxy eQTLs for rs117158921, while for rs200091415 a proxy variant (rs79208020 with D’ = 0.749 and r2 = 0.119) was identified for the decaprenyl diphosphate synthase subunit 1 (PDSS1) gene and another variant (rs72481178 with D’ = 0.592 and r2 = 0.344) for FAM238B. PDSS1 is minimally expressed in DRG (FPKM of 1.56), while no expression data was available for FAM238B. No tibial nerve eQTLs were identified for rs9846958 on chromosome 3, and the closest gene is glutamate decarboxylase like 1 (GADL1) (Additional file 1: Fig. S3D), which had minimal expression in the DRG (FPKM of 0.09).
Candidate SNP analysis
Based on prior GWAS studies examining paclitaxel-induced CIPN (Additional file 1: Table S2) [20, 25,26,27,28], we performed focused candidate gene analyses which only included variants with significant association (at suggestive genome-wide significance level of p < 10–5) between the polymorphism and CIPN (Additional file 3: Table S3). No variants were identified with P values that exceeded correction for multiple testing.
Polygenic risk scores generated from patient-reported CIPN GWAS
We examined the effect of multiple variants by calculating a polygenic risk score (PRS) comprising of SNPs with P values from the GWAS falling within identified thresholds (ranging from P > 10–10; 10–8, 10–7, 10–6, 10–5 and 1; Fig. 1C). A PRS calculated from 11 top-ranked variants (Table 1) with P values < 10–6, after adjusting for variants in linkage disequilibrium, was significantly correlated (r2 = 0.34; P = 5.36 × 10–20) with patient-reported CIPN (EORTC-QLQ-CIPN20 scores). The inclusion of a further 35 SNPs in the calculation of the PRS improved the association (r2 = 0.53; P = 1.54 × 10–35) between risk score and the patient-reported measure of CIPN (Fig. 1C).
Gene-based and pathway analyses relevant to all three measures of CIPN
In order to identify common pathways relevant across different measures of CIPN, we calculated gene-based P values using PASCAL to yield a list of loci with gene-based P values of < 0.01 derived from the GWAS of each measure of CIPN. Overlap analysis was performed to identify n = 3338 genes which were in common between the patient-reported CIPN, neurological grading scale and clinical grading scale GWASs (Fig. 2). The common gene set was subsequently analysed for enrichment of gene ontology (GO) and Reactome pathways [29]. There were 10 GO terms that were significantly over-represented (Table 4, False discovery rate < 0.05), including the glutamate receptor signalling pathway (GO:0007215, enrichment ratio 2.8, adjusted P value = 3.85 × 10–7) and axon development (GO:0061564, enrichment ratio 1.65, adjusted P value = 1.78 × 10–6). Similarly, there were ten Reactome pathways that were significantly over-represented (Table 4), including Na+/Cl− dependent neurotransmitter transporters (R-HSA-442660, enrichment ratio 4.4, adjusted P = 0.00033152) and Neuronal system (R-HSA-112316, enrichment ratio 1.9, adjusted P = 3.33 × 10–7).
Discussion
Chemotherapy-induced peripheral neurotoxicity is a significant adverse event of paclitaxel treatment that can lead to early treatment discontinuation, persistent functional disability and reduced quality of life [2, 3]. In this study, we performed a GWAS on 183 patients treated with paclitaxel to identify genetic variants associated with CIPN, as measured using multiple neuropathy outcome measures. We identified multiple SNPs with genome-wide significance associated with patient-reported neuropathy. Pathways analysis was used to identify mechanistic pathways involved in CIPN and a polygenic risk score was determined. Importantly, our findings highlight the potential role of axon development and regeneration pathways in paclitaxel-induced CIPN.
Our study identified 4 chromosomal regions (rs9846958, nearest gene GADL1 on chromosome 3; rs117158921, nearest gene AC124254.2 on chromosome 18; rs4560447, nearest gene LIMCH1 on chromosome 4; rs200091415, nearest gene FAM238B on chromosome 19) that passed genome-wide significance in the patient-reported neuropathy GWAS (P < 5 × 10–8; Table 2). Prior GWAS (E5103 (26), CALGB 40101 [25, 27, 28] and a meta-analysis of two GWAS studies (CALGB 40502 and CALGB 40101[20]) on patients treated with paclitaxel have identified a range of SNPs associated with neuropathy, but none exceeded genome-wide significance. In our study, the potential impact of top associated variants on the function of non-coding RNAs was highlighted by VEP annotation (Fig. 1B, Additional file 2: Table S1). This is consistent with results from a transcriptomic study that identified dysregulation of long non-coding RNAs and mRNAs mediating neuroinflammation and pain in the spinal cord of a rat model of paclitaxel-induced peripheral neuropathy [30].
While functional annotations have traditionally focused on known genes, thousands of disease-associated SNPs are located within intergenic regions, making it difficult to understand their association with disease phenotypes. Recent analyses found that non-coding disease associated SNPs were frequently located in or approximate to regulatory elements, such as the binding sites for CCCTC-binding factors (CTCF) and enhancer elements that act distally to promote gene expression [31]. In our annotation of the top 54 associated SNPs (Additional file 2: Table S1), 4 were located within these regulatory elements. CADD scores are based on various genomic features derived from surrounding nucleotide sequences, gene model annotations, evolutionary constraints, epigenetic marks and functional predictions [32]. We observed that 2 intergenic SNPs had CADD scores greater than 10, that is they were ranked in the top 10% of all known variants likely to be deleterious (Additional file 2: Table S1). We also note that one of the top associated SNP (rs9846958) on chromosome 3 would be considered to be an intergenic SNP and currently lacks any functional annotation, but we have indicated the closet gene to be GADL1 (Fig. 1A).
In the present study, of the 4 SNPs with genome-wide significance, the associated gene LIMCH1 was most prominently expressed in the DRG, a key region implicated in CIPN pathogenesis. LIMCH1 has been identified as a key regulator of actin-cytoskeleton remodelling, involved in cell migration [22]. Due to its role in cell migration and adhesion, LIMCH1 has been associated with worse prognosis in multiple forms of cancer [33]. While LIMCH1 has not been directly associated with nerve function, actin-cytoskeletal frameworks are critical in neuronal development, and axonal growth and actin-binding LIM domain proteins are important in axonal regeneration [34]. Another actin-binding protein LIMK2, which acts to regulate cell proliferation and migration, has also been linked to paclitaxel-induced CIPN in a prior GWAS [27].
Further, there is substantial evidence highlighting the potential role of actin cytoskeleton and axonal guidance pathways in paclitaxel-induced CIPN [35]. Comparison of differences in signalling pathways and gene co-expression between paclitaxel-treated patients with and without CIPN provided molecular evidence of the involvement of cytoskeletal and axonal morphology pathways in neuropathy development [35]. This included a suite of genes previously associated with paclitaxel-induced neuropathy, including the EPHA gene family linked to receptors for axonal grown and neural development [27, 28] and FDG4, a F-actin binding protein [29]. Of note, although no candidate variants were independently replicated in our GWAS dataset, there was some support for EPHA5 (genotyped SNP rs3605041, P = 0.0021 for TNSc-GWAS), which encodes an ephrin receptor important in neurite growth during development [7].
In further support of the importance of axonal and cytoskeletal development pathways in paclitaxel-induced CIPN, a key gene-ontology pathway of interest from our analysis across multiple outcome measures was the axon development pathway. This underscores the results of previous analyses, which have highlighted this pathway as central to paclitaxel-induced PN [7, 36]. Consistent with our findings (Table 4), differential gene expression and pathway impact analysis identified significantly perturbed cytoskeleton- and axon morphology-related signalling pathways in patients treated with paclitaxel [35]. These pathways have recently been highlighted in conjunction with their links to Ras homolog family of guanosine triphosphate hydrolase (RhoGTPase) signalling pathways relevant to axon extension and cell mobility [7]. RhoGTPases are important in sensory neuronal development and outgrowth as well as axonal regeneration [37] and are linked to paclitaxel-induced PN development, including via LIM domain proteins [37].
Although our study has identified several variants with genome-wide significance, we did not independently replicate the findings of prior studies in our dataset, given the number of candidate variants examined. However, the top replicated variant was rs9332998 from the CIPN20-GWAS, a proxy for rs4646487 within the CYP4B1 gene. The gene is part of the CYP genes set that modulate paclitaxel pharmacokinetics and similar genes have been associated with paclitaxel-induced CIPN in prior analyses [9]. Replication studies have often failed to confirm genetic associations in CIPN, potentially related to a lack of standardisation in outcome measures, with different thresholds for CIPN case identification affecting findings [9].
We utilised multiple CIPN outcome measures, including patient reported symptoms, clinical grading scale and neurological assessment. While there remains no gold standard CIPN assessment tool, evidence suggests that multimodal CIPN assessment incorporating both patient report and clinician assessment may present the most comprehensive information about neuropathy status [11]. However, only a minority of prior genetic risk factor studies have utilised patient-reported outcomes [38]. Importantly, patients typically report greater severity of symptoms than reported by clinicians [10] and this has been demonstrated to affect the identification of genetic risk factors for paclitaxel-induced CIPN [38]. Conversely, there has been criticism of relying solely on patient-reported CIPN assessment for biomarker studies, as patient report may be more variable and lack a consistent benchmark of severity compared to clinical assessment [39]. Of note, in this study, SNPs with genome-wide significance were only identified in the GWAS using patient reported CIPN. This may be related to the sensitivity of patient-reported outcomes for neuropathy but may also reflect limitations in more objective outcomes which do not always match with patient report [11].
Another factor that complicates the search for genetic variants associated with paclitaxel-induced PN is the likely polygenic inheritance, with multiple variants contributing to the risk of PN [36, 40]. It is likely that a large number of SNPs each contribute a small, additive risk to the development of paclitaxel-induced PN [36, 40]. Importantly, the use of polygenic risk scores (PRS) which aggregate the effects of multiple genetic variants across the human genome into a single score, have recently been shown to have predictive value for multiple common diseases such as breast cancer and diabetes [41]. Further, the integration of genetic information with non-genetic risk factors has been demonstrated to enhance the sensitivity and specificity of PRS as a clinical tool [42]. In our dataset, a PRS calculated from 46 SNPs was highly correlated with patient-reported CIPN (Fig. 1C). Our PRS differs from scores calculated for idiopathic neurodegenerative diseases such as Alzheimer’s disease [43] which typically require > 100,000 SNPs and have poorer predictive values with r2 < 0.1. This may reflect the fact that pharmacogenomic variants typically have stronger genetic effects compared with common disease-associated variants [44]. We also note that our PRS is calculated from patients of European descent, and validation of our PRS by other investigators should involve controlling for population stratification. This is especially important as the rate of severe CIPN may vary by ethnicity [9, 25]. Nonetheless, such an approach is likely to be beneficial for the prediction of CIPN and should form the basis for future genetic analyses of CIPN.
Strengths and limitations
This study has identified several variants with genome-wide significance linked to paclitaxel-induced peripheral neuropathy. A strength of the study was the inclusion of multiple neuropathy assessment tools, including validated patient reported outcomes. However, a limitation of our GWAS is the sample size, which may affect statistical power. Our findings should be replicated in larger datasets, preferably with diverse populations. In addition, our sample included multiple treatment protocols and cancer types, heterogeneity which may affect the generalizability of results to specific cohorts. Accordingly, our loci and PRS require validation and replication in independent datasets, preferably with compatible CIPN outcome measures. It should be noted that lack of standardization in CIPN outcome measures across studies and in particular in large-scale clinical trials of neurotoxic agents has limited the ability for data from different studies to be meaningfully combined. Hopefully efforts to standardize outcome measures for CIPN will assist towards this aim.
Conclusions
In conclusion, we have identified novel genetic loci associated with patient-reported paclitaxel- induced peripheral neuropathy and these findings provide further evidence for the involvement of axon development pathways in paclitaxel-induced CIPN. Our study highlights the importance of appropriate and patient-relevant CIPN outcome measures in defining the CIPN phenotype. In total, this study highlights the polygenic nature of CIPN risk, as definition of polygenic patterns of inheritance will be critical to ultimately enable genetic risk factors to become useful tools to predict patient risk in the clinic and improve patient quality of life following paclitaxel treatment.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- BMI:
-
Body mass index
- CADD:
-
Combined Annotation Dependent Depletion
- CIPN:
-
Chemotherapy induced peripheral neuropathy
- DRG:
-
Dorsal root ganglia
- EORTC QLQ-CIPN20:
-
European Organization for Research and Treatment of Cancer Quality of Life Questionnaire-Chemotherapy-Induced Peripheral Neuropathy
- eQTL:
-
Expression quantitative trait loci
- GTEx:
-
Genotype-Tissue Expression
- GWAS:
-
Genome wide association study
- NCI-CTCAE:
-
National Cancer Institute Common Terminology Criteria for Adverse Events
- PRS:
-
Polygenic risk score
- RhoGTPase:
-
Guanosine triphosphate hydrolase
- SNP:
-
Single nucleotide polymorphism
- TNSc:
-
Total Neuropathy Score–clinical version
- Q-Q plots:
-
Quantile–quantile plots
- VEP:
-
Variant Effect Predictor
References
Weaver BA. How Taxol/paclitaxel kills cancer cells. Mol Biol Cell. 2014;25(18):2677–81.
Park SB, Goldstein D, Krishnan AV, Lin CS, Friedlander ML, Cassidy J, et al. Chemotherapy-induced peripheral neurotoxicity: a critical analysis. CA Cancer J Clin. 2013;63(6):419–37.
Battaglini E, Goldstein D, Grimison P, McCullough S, Mendoza-Jones P, Park SB. Chemotherapy-induced peripheral neurotoxicity in cancer survivors: predictors of long-term patient outcomes. J Natl Compr Canc Netw. 2021;19(7):821–8.
Hertz DL, Childs DS, Park SB, Faithfull S, Ke Y, Ali NT, et al. Patient-centric decision framework for treatment alterations in patients with Chemotherapy-induced Peripheral Neuropathy (CIPN). Cancer Treat Rev. 2021;99: 102241.
Loprinzi CL, Lacchetti C, Bleeker J, Cavaletti G, Chauhan C, Hertz DL, et al. Prevention and management of chemotherapy-induced peripheral neuropathy in survivors of adult cancers: ASCO guideline update. J Clin Oncol. 2020;38(28):3325–48.
LaPointe NE, Morfini G, Brady ST, Feinstein SC, Wilson L, Jordan MA. Effects of eribulin, vincristine, paclitaxel and ixabepilone on fast axonal transport and kinesin-1 driven microtubule gliding: implications for chemotherapy-induced peripheral neuropathy. Neurotoxicology. 2013;37:231–9.
Chua KC, El-Haj N, Priotti J, Kroetz DL. Mechanistic insights into the pathogenesis of microtubule-targeting agent-induced peripheral neuropathy from pharmacogenetic and functional studies. Basic Clin Pharmacol Toxicol. 2021. https://doi.org/10.1111/bcpt.13654.
Staff NP, Fehrenbacher JC, Caillaud M, Damaj MI, Segal RA, Rieger S. Pathogenesis of paclitaxel-induced peripheral neuropathy: a current review of in vitro and in vivo findings using rodent and human model systems. Exp Neurol. 2020;324: 113121.
Cliff J, Jorgensen AL, Lord R, Azam F, Cossar L, Carr DF, et al. The molecular genetics of chemotherapy-induced peripheral neuropathy: a systematic review and meta-analysis. Crit Rev Oncol Hematol. 2017;120:127–40.
Tan AC, McCrary JM, Park SB, Trinh T, Goldstein D. Chemotherapy-induced peripheral neuropathy-patient-reported outcomes compared with NCI-CTCAE grade. Support Care Cancer. 2019;27(12):4771–7.
Alberti P, Rossi E, Cornblath DR, Merkies IS, Postma TJ, Frigeni B, et al. Physician-assessed and patient-reported outcome measures in chemotherapy-induced sensory peripheral neurotoxicity: two sides of the same coin. Ann Oncol. 2014;25(1):257–64.
Cavaletti G, Frigeni B, Lanzani F, Piatti M, Rota S, Briani C, et al. The Total Neuropathy Score as an assessment tool for grading the course of chemotherapy-induced peripheral neurotoxicity: comparison with the National Cancer Institute-Common Toxicity Scale. J Peripher Nerv Syst JPNS. 2007;12(3):210–5.
Postma TJ, Aaronson NK, Heimans JJ, Muller MJ, Hildebrand JG, Delattre JY, et al. The development of an EORTC quality of life questionnaire to assess chemotherapy-induced peripheral neuropathy: the QLQ-CIPN20. Eur J Cancer. 2005;41(8):1135–9.
Boughton AP, Welch RP, Flickinger M, VandeHaar P, Taliun D, Abecasis GR, et al. LocusZoom.js: interactive and embeddable visualization of genetic association study results. Bioinformatics. 2021;37(18):3017–8.
Choi SW, Mak TS, O’Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc. 2020;15(9):2759–72.
Myers TA, Chanock SJ, Machiela MJ. LDlinkR: an R package for rapidly calculating linkage disequilibrium statistics in diverse populations. Front Genet. 2020;11:157.
Lamparter D, Marbach D, Rueedi R, Kutalik Z, Bergmann S. Fast and rigorous computation of gene and pathway scores from SNP-based summary statistics. PLoS Comput Biol. 2016;12(1): e1004714.
Alonso-Gonzalez A, Calaza M, Rodriguez-Fontenla C, Carracedo A. Novel gene-based analysis of ASD GWAS: insight Into the biological role of associated genes. Front Genet. 2019;10:733.
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The ensembl variant effect predictor. Genome Biol. 2016;17(1):122.
Chua KC, Xiong C, Ho C, Mushiroda T, Jiang C, Mulkey F, et al. Genomewide meta-analysis validates a role for S1PR1 in microtubule targeting agent-induced sensory peripheral neuropathy. Clin Pharmacol Ther. 2020;108(3):625–34.
Flegel C, Schöbel N, Altmüller J, Becker C, Tannapfel A, Hatt H, et al. RNA-seq analysis of human trigeminal and dorsal root ganglia with a focus on chemoreceptors. PLoS ONE. 2015;10(6): e0128951.
Lin YH, Zhen YY, Chien KY, Lee IC, Lin WC, Chen MY, et al. LIMCH1 regulates nonmuscle myosin-II activity and suppresses cell migration. Mol Biol Cell. 2017;28(8):1054–65.
Lonsdale J, Thomas J, Salvatore M, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–5.
Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–7.
Komatsu M, Wheeler HE, Chung S, Low SK, Wing C, Delaney SM, et al. Pharmacoethnicity in paclitaxel-induced sensory peripheral neuropathy. Clin Cancer Res. 2015;21(19):4337–46.
Schneider BP, Li L, Radovich M, Shen F, Miller KD, Flockhart DA, et al. Genome-wide association studies for taxane-induced peripheral neuropathy in ECOG-5103 and ECOG-1199. Clin Cancer Res. 2015;21(22):5082–91.
. LeandroGarcía LJ, IngladaPérez L, Pita G, Hjerpe E, Leskelä S, Jara C, et al. Genome-wide association study identifies ephrin type A receptors implicated in paclitaxel induced peripheral sensory neuropathy. J Med Genet. 2013;50(9):599–605.
Baldwin RM, Owzar K, Zembutsu H, Chhibber A, Kubo M, Jiang C, et al. A genome-wide association study identifies novel loci for paclitaxel-induced sensory peripheral neuropathy in CALGB 40101. Clin Cancer Res. 2012;18(18):5099–109.
Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45(W1):W130–7.
Li Y, Yin C, Liu B, Nie H, Wang J, Zeng D, et al. Transcriptome profiling of long noncoding RNAs and mRNAs in spinal cord of a rat model of paclitaxel-induced peripheral neuropathy identifies potential mechanisms mediating neuroinflammation and pain. J Neuroinflammation. 2021;18(1):48.
Corradin O, Scacheri PC. Enhancer variants: evaluating functions in common disease. Genome Med. 2014;6(10):85.
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–94.
Cao H, Zhao J, Chen Z, Sun W, Ruan K, Zhou J, et al. Loss of LIMCH1 predicts poor prognosis in patients with surgically resected Lung Adenocarcinoma: a study based on Immunohistochemical Analysis and Bioinformatics. J Cancer. 2021;12(1):181–9.
Levin E, Leibinger M, Gobrecht P, Hilla A, Andreadaki A, Fischer D. Muscle LIM protein is expressed in the injured adult CNS and promotes axon regeneration. Cell Rep. 2019;26(4):1021-32.e6.
Kober KM, Schumacher M, Conley YP, Topp K, Mazor M, Hammer MJ, et al. Signaling pathways and gene co-expression modules associated with cytoskeleton and axon morphology in breast cancer survivors with chronic paclitaxel-induced peripheral neuropathy. Mol Pain. 2019;15:1744806919878088.
Chhibber A, Mefford J, Stahl EA, Pendergrass SA, Baldwin RM, Owzar K, et al. Polygenic inheritance of paclitaxel-induced sensory peripheral neuropathy driven by axon outgrowth gene sets in CALGB 40101 (Alliance). Pharmacogenomics J. 2014;14(4):336–42.
Kalpachidou T, Spiecker L, Kress M, Quarta S. Rho GTPases in the physiology and pathophysiology of peripheral sensory neurons. Cells. 2019. https://doi.org/10.3390/cells8060591.
Park SB, Kwok JB, Asher R, Lee CK, Beale P, Selle F, et al. Clinical and genetic predictors of paclitaxel neurotoxicity based on patient- versus clinician-reported incidence and severity of neurotoxicity in the ICON7 trial. Ann Oncol. 2017;28(11):2733–40.
Hertz DL. Concerns regarding use of patient-reported outcomes in biomarker studies of chemotherapy-induced peripheral neuropathy. Pharmacogenomics J. 2019;19(5):411–6.
Wheeler HE, Gamazon ER, Wing C, Njiaju UO, Njoku C, Baldwin RM, et al. Integration of cell line and clinical trial genome-wide analyses supports a polygenic architecture of Paclitaxel-induced sensory peripheral neuropathy. Clin Cancer Res. 2013;19(2):491–9.
Crouch DJM, Bodmer WF. Polygenic inheritance, GWAS, polygenic risk scores, and the search for functional variants. Proc Natl Acad Sci USA. 2020;117(32):18924–33.
Lambert SA, Abraham G, Inouye M. Towards clinical utility of polygenic risk scores. Hum Mol Genet. 2019;28(R2):R133–42.
Chaudhury S, Brookes KJ, Patel T, Fallows A, Guetta-Baranes T, Turton JC, et al. Alzheimer’s disease polygenic risk score as a predictor of conversion from mild-cognitive impairment. Transl Psychiatry. 2019;9(1):154.
Maranville JC, Cox NJ. Pharmacogenomic variants have larger effect sizes than genetic variants associated with other dichotomous complex traits. Pharmacogenomics J. 2016;16(4):388–92.
Acknowledgements
We thank all participants of the study.
Funding
This study was supported by a Cancer Institute NSW Program Grant (14/TPG/1-05) and a National Health and Medical Research Council of Australia (NHMRC) Project Grant (GNT1080521). SBP is supported by a NHMRC Career Development Fellowship (GNT1148595). JBK and BG are supported by NHMRC Project Grant (GNT1163249).
Author information
Authors and Affiliations
Contributions
DG, SBP and JBK conceived and/or designed the work that led to the submission; KH, BG, HCT, TL, MH, CRL, JGL, GMT, SBP and JBK acquired data, and/or played an important role in interpreting the results. KH, DG, HCT, TL, MH, MLF, CRL, JGL, GMT, BG, SBP and JBK drafted or revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethics approval was granted by the Sydney Local Health District and South-Eastern Sydney Local Health District Human Research Ethics Committees. All patients provided written informed consent to participate.
Consent for publication
Not applicable.
Competing interests
BG is a director of Pacific Analytics PTY LTD & SMRTR PTY LTD, Australia. The authors declare that they have no conflicts of interest with the contents of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S2. Prior Genome-wide association studies examining predictors of CIPN in paclitaxel-treated patients. Figure S1. Distribution of CIPN severity in the patient cohort assessed using NCI-CTCAE clinical grading scale. Figure S2. (A) Q-Q and (B) Manhattan plots for GWAS of the three measures of CIPN including clinical grading scale (NCI), neurological grading scale (TNSc) and patient report (EORTC). Figure S3: Genetic loci corresponding to top associated SNPs identified by GWAS of patient reported EORTC-QLQ-CIPN20 CIPN visualized using LocusZoom.
Additional file 2:
Table S1. Annotation of SNPs using Ensembl Variant Predictor platform.
Additional file 3:
Table S3. Candidate SNPs derived from previously reported associations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hooshmand, K., Goldstein, D., Timmins, H.C. et al. Polygenic risk of paclitaxel-induced peripheral neuropathy: a genome-wide association study. J Transl Med 20, 564 (2022). https://doi.org/10.1186/s12967-022-03754-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-022-03754-4