Abstract
Background
Plasmodium vivax contributes to over 70% malaria burden in Pakistan, but limited data exists on various aspects including genetic diversity of the parasite as compared to other parts of the world. Since the information about the genetic diversity of P. vivax assists to understand the population dynamics of the parasite, the current study was designed to understand population divergence of P. vivax in Pakistan using circumsporozoite protein (pvcsp) and merozoite surface protein-1 (pvmsp-1) genes as molecular markers.
Methods
The PCR for pvcsp and pvmsp-1 genes was carried out for 150 P. vivax isolates, followed by DNA sequencing of 35 and 30, respectively. Genetic diversity and polymorphism were analysed using ChromasPro, ClustalW, MEGA7, DnaSP v.5 and WebLogo programs.
Results
The PCR for pvcsp and pvmsp-1 genes was carried out for 150 P. vivax isolates and resulting the PCR products of 1100 bp for pvcsp and ~ 400 bp for pvmsp-1 genes, respectively. In the central-repeat region (CRR) of pvcsp gene, sequences comprised of four variable repeats of PRMs, out of which GDRADGQPA (PRM1), GDRAAGQPA (PRM2) were more extensively dispersed among the P. vivax isolates. Partial sequences (~ 400 bp) of block 2 of pvmsp-1 gene depicted high level of diversity.
Conclusion
The results revealed the polymorphism and genetic diversity especially at the CRR of pvcsp and block 2 of pvmsp-1 genes, respectively. The base-line data presented here warrants future studies to investigate more into the genetic diversity of P. vivax with large sample size from across the country for better understanding of population dynamics of P. vivax that will help to control malaria at individual and community level.
Similar content being viewed by others
Background
Malaria remains a serious infectious disease of public health importance and one of the leading causes of morbidity and mortality worldwide [1]. Globally, around 42% population is at risk with almost 228 million cases of malaria with 0.4 million reported deaths in 2018 based on the data collected from 91 countries [2, 3]. Among the five Plasmodium species, Plasmodium vivax contributes around 70% malaria cases in Pakistan [4] with variable severity [5,6,7]. To circumvent the parasite load, there is a need to investigate the population structure, genetic diversity [8], parasite typing of the local P. vivax species, pattern of antigenic variations and drug resistance [9, 10]. It would lead to interruption of transmission cycle of the parasite in human host in endemic areas. Despite multiple academic research ventures, scanty data is available regarding the diverse genetic make-up of P. vivax in Pakistan [5, 6].
Genetic sequences of pvcsp and pvmsp-1 are being used to understand the genetic diversity [11,12,13]. The pvcsp is a highly immunogenic sporozoite surface protein, hence a good vaccine candidate, is encoded by single copy gene [8] and comprises of a central repeat domain that varies across Plasmodium species having two non-repetitive domains at N- and C-terminals [14, 15]. The varying degree of number of peptides in the central repeat region reveals three variants of P. vivax namely VK210, VK247, P. vivax-like [16, 17]. Across the globe, these variants exhibit certain spatial predispositions. With a GDRA(A/D)GQPA amino acid repetition, VK210 strain dominates in the endemic region [18, 19]. Originating in Thailand, the VK247 strain is mostly reported from the areas where mixed infections are prominent [20]. VK247 depicts ANGAGNQPG amino acid repeat in the central region [20, 21]. In pvcsp, polymorphism is reported to be limited in central tandem repeat among the isolates from the regions of sub-Saharan Africa, China, Sri Lanka, Sudan and India [8,9,10, 13, 14, 22]. The pvmsp-1 is expressed on the surface of the blood-stage parasite [23], a large gene, covers conserved and polymorphic regions [24] and has mosaic organization with 13 regions of variable blocks [25]. The three main regions of sequence divergence are block 2 (F1 region), 6–8 (F2 region) and 10 (F3 region) [26]. In the representative blocks, the genetically distinct pvmsp-1 populations within the regions and polymorphism can be detected through PCR [5, 27]. Selective pressure of the host immune maintains the diversity of pvmsp-1 gene, however, immunogenic properties can be affected by single-point mutation [28, 29]. Despite P. vivax contributing to 88% of malaria burden in Pakistan [5,6,7], data regarding genetic diversity of this key circulating species is lacking. The present study was designed to investigate the genetic diversity of P. vivax in Potohar region of Pakistan exploiting pvcsp and pvmsp-1 genes as molecular markers. The understanding of sequence diversity in pvcsp would contribute to comprehend the population dynamics and transmission patten of the parasite in this region.
Methods
Study area
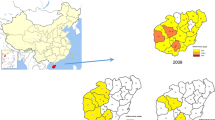
The study was conducted in Islamabad and Rawalpindi districts of the Punjab with longitudes 72°45′ and 73°30′ E and latitudes 33°30′ and 33°50′ N [30] (Fig. 1). The climate in the study region ranges from showery warm to chilly dry wintry with the attributes of the semi-arid region of Pakistan. The monsoon rains typically start in June, get peak in August, and finish by September. The rainfall is between 620 and 1200 mm per year. The weather and geographical settings of this region are favorable for the mosquito breeding with highest frequency reported in Rawalpindi (25.5%) and lowest in Chakwal (15.9%) [31].

available at https://malariaatlas.org/. The estimated sites of the study sites described here are showed with red star: Rawalpindi and Islamabad
Map of P. vivax prevalence in EMRO region and Pakistan. The map showing all age clinical cases of P. vivax from year 2000–2017 of EMRO region and Pakistan. These maps were originally created by the Malaria Atlas Project, University of Oxford
Sample collection
Blood samples (n = 150) were collected during malaria transmission season (April and October, 2019) from Pakistan Institute of Medical Sciences (PIMS), Islamabad and Rawalpindi General Hospital, Rawalpindi, Pakistan. Venous blood (5 mL) was collected in ethylene diamine tetra-acetic acid (EDTA) (BD, USA) vacutainers from malaria patients. The samples were collected from the patients fulfilling the inclusion criteria of clinical signs and symptoms (fever, chills, headache, sweats, fatigue, nausea and vomiting). Initially malaria patients were screened by microscopic examination of Giemsa-stained thin and thick blood smears to ensure the P. vivax parasites and exclude samples with presence of mixed Plasmodium species infections (mixed P. vivax and P. falciparum). Blood samples were stored at − 20 °C for a month until further analysis was carried out.
DNA extraction and PCR amplification of pvcsp and pvmsp-1 genes
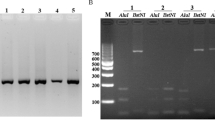
Genomic DNA from 150 malarial blood samples were extracted by using standard phenol–chloroform method [32]. PCR primers for pvcsp and pvmsp-1 genes were designed at Geneious software by using reference sequence of P. vivax (AB539044 and GQ890906). The primers for pvcsp gene were F: 5′-GGCCATAAATTTAAATGGAG-3′and R: 5′-ATGCTAGGACTAACAATATG-3′. The PCR conditions were as follows: pre-denaturation at 94 °C for 10 min followed by 35 cycles of annealing at 52 °C for1 min and extension at 72 °C for 1 min, followed by final extension at 72 °C for 10 min. The PCR of pvmsp-1 gene was performed with primers F: 5′-ACATCATTAAGGACCCATACAAG 3′ and R: 5′ GCAATTTCTTTACAGTGATCTCG-3′ with similar PCR cycling conditions except that the annealing was at 56 °C for 1 min. The PCR reactions were carried out in a 25 μL reaction mixture comprising of 2μL DNA template, 0.5 mM dNTPs, 1X PCR reaction buffer (SolarBio Life Sciences, China), 0.2 mM of each primer (BGI, China), 2.5 mM MgCl2 and 1 unit of Taq DNA polymerase (BLIRT, Poland). The PCR products were visualized using 1% agarose gel (ThermoFisher, USA) stained with ethidium bromide (SolarBio Life Sciences, China) and visualized under UV-trans illuminator (ThermoFisher, USA).
Sanger sequencing and analysis
All 150 malaria amplified genomic DNA samples showed the band size of 1100 bp for pvcsp gene and ~ 400 bp for partial sequence of pvmsp-1 gene. As no variation existed in band sizes of 150 malaria samples for both genes, only 35 and 30 amplified PCR products of both pvcsp and pvmsp-1 genes were selected randomly and purified, respectively by QIAquick PCR Purification Kit (Qiagen, Germany according to manufacturer’s protocol). These purified samples were then sent to Beijing Genomics Institute (BGI), China for Sanger sequencing. DNA sequences of both pvcsp and pvmsp-1 genes were read and assembled on both upstream and downstream ends for 35 sequences of pvcsp gene and 30 sequences of pvmsp-1 gene. The sequences were then analysed by using ChromasPro (version 1.5) software (http://technelysium.com.au/wp/chromaspro/) and Bio Edit alignment editor (version 7.2) (https://bioedit.software.informer.com/7.2/). The sequenced samples were validated by BlastN (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and pvcsp repeat types (VK210/VK247) was analysed by BLAST. The alignment of top hit resulted sequences were done by ClustalW (https://www.genome.jp/tools-bin/clustalw). The number of haplotypes (H), nucleotide diversity (π) and haplotype diversity (Hd) were calculated by DNAsp v5 [33]. The evolutionary relationships of the both genes were established, and evolutionary tree were constructed by the neighbour-joining method using Molecular Evolutionary Genetics Analysis (MEGA 7.0) software [34]. The neutral theory of natural selection and population bottlenecks were also measured by Tajima’s D, Fu and Li’s D*, and Fu and Li’s F* tests using DNAsp v5 [33]. The negative value of Tajima’s D, Fu and Li’s D* indicates an excess of rare alleles that might results from selective sweep. A plot was constructed to look for polymorphic patterns of the N- and C-terminal in Pakistani pvmsp-1 gene by using the WebLogo program (https://weblogo.berkeley.edu/logo.cgi). The nucleotide sequences of pvcsp and pvmsp-1 genes obtained from this study were submitted in NCBI database under accession number MT222296 to MT222330 and MT303819 to MT303848, respectively.
Results
In the present study, a total of 150 blood samples were collected from P. vivax malaria infected patients from the hospitals of twin cities of Islamabad and Rawalpindi. The blood samples were verified by microscopic examination to ensure the P. vivax parasites and excluded the samples for presence of mixed Plasmodium species infections. The PCR product of 1100 bp for pvcsp gene and ~ 400 bp for partial sequence of block 2 for pvmsp-1 gene.
Sequence analysis of pvcsp gene
Multiple sequence alignment of the translated nucleotide sequences was carried out for the analysis of polymorphisms in the pre-, post- and central repeats of the pvcsp gene. The top hits for pvcsp gene were extracted from GenBank protein database using Blastp and one of the sequences of pvcsp gene of Iranian isolate was retrieved and used as reference sequence (KT588208.1). The multiple sequence alignment of extracted amino acid sequences was performed using ClustalW. When compared with the reference sequence (KT588208.1), the sequence analysis of pvcsp gene showed the VK210 and VK247 variant types infection. The pvcsp gene sequence analysis revealed that majority (92%; 32/35) of the P. vivax isolates were of VK210 variant and only 3 isolates were found to be VK247 type. All the pvcsp gene-based P. vivax variants started with the same pre-repeat sequence (KLKQP region I). In the central-repeat region (CRR), the VK210 sequences comprised of variable repeats of PRMs, GDRADGQPA (PRM1), GDRAAGQPA (PRM2) which were found in all the isolates. It was followed through two conserved post-repeat sequence GNGAGGQAA (PRM3) and GGNAANK (PRM4) and one post-repeat insert i.e., KAEDA region. The one-copy repeat region of GGNA was found after the CRR in all the analysed sequences. The frequency of peptide repeat motifs (PRMs) in the central repeat region (CRR) of pvcsp has been summarized in Fig. 2. The observed non-synonymous substitution based on diverse types of repetition in allotypes (RATs), which leads to different PRMs are mentioned in Table 1.
Pvcsp CRR based genetic population structure
The population genetic structure based on the pvcsp CRR of the P. vivax isolates was analysed and compared with pvcsp isolates of neighboring countries Iran, India and Myanmar. The haplotype (gene) diversity of pvcsp was categorized into fifteen distinct haplotypes with an estimated Hd of 0.547 and ten distinct haplotypes with an estimated Hd of 0.345 in Pakistani and Iranian pvcsp samples respectively. The values for Tajima’s D, Fu and Li’s D* and F* tests are given in Table 2 for the pvcsp variants from Pakistan, Iran, India and Myanmar. The Fu and Li’s D* and F* values for CRR region was also positive suggested that the CRR region of pvcsp population of Pakistan was under positive natural selection. The nucleotide diversity in pvcsp population of Pakistan, Iran, India and Myanmar were highly significant (P < 0.05) as compared to haplotype diversity which was significant (P < 0.05) in Pakistan and Iran and non-significant (P > 0.05) in India and Myanmar. The pvcsp population from India and Iran showed high nucleotide diversity but values from Myanmar of pvcsp population were negative, suggesting negative selection. The values of the Tajima’s D, Fu and Li’s D* and F* values for CRR region was also positive for pvcsp population of Iran and India as shown in Table 2.
Phylogenetic analysis of pvcsp gene
A phylogenetic tree drawn from the sequence findings of pvcsp gene is presented in Fig. 3. Two separate clades can be inferred from the tree; one having VK210 variant type while the other has VK247 variant type of pvcsp gene. Four sub-clusters of VK210 and VK247 can be distinguished in the leading clade. The associated taxa were clustered together and shown after the branches with the branch length as of the evolutionary distances used to calculate the phylogenetic tree. The evolutionary distance was computed by the p-distance method and analysed using 55 nucleotide sequences. All the positions that had gaps or missing data were discarded. VK210 strain sequences from Pakistan showed 54% identity with pvcsp sequences from countries such as Iran, Greece, India, USA, Sri Lanka, Australia, Vanuatu and 100% with Myanmar, whereas the sequences of VK247 strains from Pakistan showed 100% identity with pvcsp sequences from Iran, Columbia, Vanuatu, USA, India and Korea.
Sequence analysis of pvmsp-1 gene
The top hits for pvmsp-1 gene were extracted from GenBank protein database using Blastp and one of the sequences of pvmsp-1 gene of Iranian isolate was retrieved and used as reference sequence (KX697612.1). Sequence of pvmsp-1 gene was compared with reference sequence KX697612.1 of Iranian P. vivax strain. It revealed that pvmsp-1 gene sequences of 30 samples were corresponding to partial sequence of block 2 of pvmsp-1 gene. Overall, 13 single nucleotide polymorphisms (SNPs) were found amongst 30 sequences with an average π value of 0.00143 in block 2 of pvmsp-1 gene. The average conserved sequence between Pakistani and reference Iranian pvmsp-1 gene was C: 0.835 indicating that sequences have remained relatively unchanged with close evolutionary relationship. Overall genetic polymorphisms of the pvmsp-1 population were analyzed as shown in Fig. 4. The low frequencies of uneven amino acid changes were identified at block 2 of the pvmsp-1 gene. The significant variation was observed from amino acid position K55N to M78T/N showing uneven and low frequencies with less conserved sequence of pvmsp-1.
Pvmsp-1 N-terminal based genetic population structure
Population genetic structure based on the N-terminal of pvmsp-1 gene of the P. vivax isolates was analysed and compared with isolates of neighboring country Iran as shown in Table 2. The haplotype diversity of pvmsp-1 gene was comparable between two countries ranging from 0.012 to 0.989. Adding to this, Tajima’s D, Fu and Li’s D* and F* tests also accepted occurrence of a neutral model of polymorphism with values for Fu and Li’s D* and Fu and Li’s F* are given in Table 2 for the pvmsp-1 variants from Pakistan and Iran. The Fu and Li’s D* and F* values for pvmsp-1 population were also positive. The results of pvmsp-1 population of Pakistan also indicated that positive natural selection may occur in the region. The overall nucleotide and haplotype diversity were 0.00162 ± 0.0000026 and 0.012 ± 0.00014 respectively. The nucleotide diversity in pvmsp-1 population of Pakistan, Iran and India were highly significant (P < 0.05) as compared to haplotype diversity, which was significant (P < 0.05) in Pakistan and Iran. The effect of natural selection was estimated by the Tajima’s D which was 1.67790 (P > 0.10).
Phylogenetic analysis of pvmsp-1 gene
Based on sequence of pvmsp-1 gene, a phylogenetic tree was constructed (Fig. 5). Two distinct clades can be inferred from the tree. The first is divided further into three sub-clades and contains Pakistani isolates and isolates belong to East Africa, Thailand, Mexico, India and USA with these isolates having 16 to 95% identity with sequences from Pakistani pvmsp-1 population. Second clade is further divided into two sub-clades having Turkey, Iran, Korea and Southern Mexico isolates in addition to sequences of Pakistani isolates with 62–94% sequence identity.
Discussion
Malaria is one of the major public health concerns with limited data in Pakistan [35]. As Pakistan shares border with malaria endemic countries like Iran, India and Afghanistan, the human migration across the border is inevitable possibly facilitating substantial cross-border transmission of malaria. The resultant recombination leads to genetic diversity and affects the frequency of new alleles in the parasite population [36, 37]. There are limited studies from Pakistan which have analysed the diversity of local P. vivax in detail [5, 6, 24, 36]. pvcsp and pvmsp-1 are among the other important genetic markers used by the researchers to understand population structure and evolutionary dynamics from different geographical regions [5, 26]. In the present study, the genetic polymorphism in the pvcsp and pvmsp-1 genes were studied in detail. The results of the study support the findings of the previously published data showing that VK210 strain is predominant type with the prevalence rate ranging between 56–100% in Iran [20, 24], Myanmar [15, 40], Brazil [39], India [14, 49], Thailand [41, 42], Azerbaijan [19], China [13, 42] and Mexico [43]. There are only few malaria endemic areas where VK247 isolates are commonly present [39].
The analysis of translated nucleotide sequences suggested that GDRADGQPA (PRM1) and GDRAAGQPA (PRM2) are two major PRMs. Earlier studies also reported the dominance of the two prime PRMs in the clinical isolates [8, 9, 14]. All these isolates were composed of similar pre-repeat sequence (KLKQP) region and conserved post-repeat sequence GGNAANK (PRM4) present as a last section in all of the VK210 isolates as aforementioned in studies from India, Iran and Sri Lanka [9, 14, 20]. Another peptide repeat motif GNGAGGQAA (PRM3) was found at lower frequency (0.6%) in the isolates. The variations exist in the amino acid and nucleotide sequences of the Plasmodium antigens due to variations in the repeat unit numbers which is indicative of the natural selection pressure by the host immune system [14, 44]. The arrangement of the main PRM1 and PRM2 factors leads to 15 different haplotypes of pvcsp. The analysis of CRR region of Pakistani pvcsp isolates is indicative of positive selection when compared with pvcsp isolates of Iran and India. The negative Tajima’s D values of Myanmar pvcsp population imply purifying negative selection [15]. The evidence from the previous studies has reported that the arrangements and numbers of PRMs in CRR are indicative of occurrence of phenomenon of natural selection on the pvcsp isolates [15, 45, 46].
The pvmsp-1 is one of the most promising vaccine candidates and is available for antigenic and genetic variation studies of P. vivax populations [47]. In this study, partial sequence (~ 400 bp) at block 2 of pvmsp-1 gene has depicted a high-level of diversity, which is in concordance with what has already been observed in neighboring country Iran [24], and also in previous study from Pakistan [5]. In the northwestern region of Thailand, a high degree of mutational variety was observed in pvmsp-1 genes of P. vivax isolates [29, 46]. The pvmsp-1 population of Pakistan has also indicated positive natural selection when compared with pvmsp-1 isolates of Iran and India. The significantly positive values of Fu and Li’s D* and F* tests may be the result of balancing the selection and population bottlenecks. The sequence diversity of the population is best studied by the intragenic recombination of pvmsp-1 gene where the allelic recombination frequencies may aid as a character reference for understanding the parasitic population structure [28]. Kibria et al. [26] also indicated a high genetic diversity of pvmsp-1 gene which undergoes selective pressure for the existence and spread of the parasite. Similar pattern of genetic diversity was observed in Pfmsp-1 gene of P. falciparum populations in Pakistan [36]. The pvmsp-1 gene sequences were useful in distinguishing the two central localities of origin in terms of geography as well as helping them to group in two different clades. These biological groups are further subdivided into different clusters according to their geographic origin [47]. The polymorphic nature of MSP-1 markers can be used to differentiate between reemergence and reinfections and to determine ultimate shifts in population dynamics of parasites [48].
Other studies carried out in India and Brazil have suggested that the mode of evolution in pvcsp gene can lead to cohort of variants that can elude the host immune response under the effect of both mitotic recombination and positive selection of new variants of P. vivax [14, 17, 49]. Therefore, it is safe to assume that the wide variety of P. vivax may be interrelated with multiple other variables including, but not limited to, genetic and biological characteristics, immunity of the host and the displacement of individuals within the boundaries of the endemic areas. Furthermore, the spread of P. vivax infections is also reinforced by relapse and early gametocytaemia, which in turn sustains local diversity, paving way for a more efficient transmission to the vector mosquitoes [13, 50]. The results revealed a broad range of genetic variety of pvcsp and pvmsp-1 genes in P. vivax population. The localities of the study areas from where samples were collected are inhabited by various ethnicities which suggested that the migration of people may carry diverse parasite entities that increase the variety of the gene-pool. The individuals infected by the disease might carry various clones which may recombine during the sexual stage of the mosquito leading to the production of offspring with new pvcsp genotypes. This prevalent phenomenon could strengthen the introduction of new strains of P. vivax into the regions where conditions for malaria transmission are conducive [13, 26].
Conclusion
The pvcsp and pvmsp-1 genes have been extensively studies to understand the genetic diversity of P. vivax population globally. This study highlights the genetic characterization of P. vivax isolates from Pakistan and results revealed the polymorphism and genetic diversity especially at the CRR of pvcsp and block 2 of pvmsp-1 genes, respectively. The results specify the effect of natural selection pressure on the malaria parasite for its survival and transmission. The base-line data presented here warrants further studies to investigate more into the genetic diversity of P. vivax with large sample size from across the country. It will help in better understanding of population dynamics of P. vivax as well as unstable reemergence of the infection due to population dynamics of parasite that will help to control malaria at individual and community level.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Ghanchi NK, Khan MH, Arain MA, Zubairi M, Raheem A, Khan MA, et al. Hematological profile and gametocyte carriage in malaria patients from Southern Pakistan. Cureus. 2019;11:e4256.
WHO. World malaria report 2020. Geneva, World Health Organization. https://www.who.int/news-room/fact-sheets/detail/malaria
Bettencourt P. Current challenges in the identification of pre-erythrocytic malaria vaccine candidate antigens. Front Immunol. 2020;11:190.
Report PMA. Malaria Control Programme. Pakistan: Directorate of Malaria Control; 2019.
Raza A, Ghanchi NK, Thaver AM, Jafri S, Beg MA. Genetic diversity of Plasmodium vivax clinical isolates from southern Pakistan using pvcsp and pvmsp1 genetic markers. Malar J. 2013;12:16.
Khan SN, Khan A, Khan S, Ayaz S, Attaullah S, Khan J, et al. PCR/RFLP-based analysis of genetically distinct Plasmodium vivax population of pvmsp-3α and pvmsp-3β genes in Pakistan. Malar J. 2014;13:355.
Ahmad T. Plasmodium vivax and Plasmodium falciparum are common malaria species in Pakistan. Biomed Res Ther. 2016;3:1–7.
Zhang LL, Yao LN, Chen HL, Lu QY, Ruan W. Genetic diversity analysis of Pvcsp and its application in tracking of Plasmodium vivax. Exp Parasitol. 2018;188:26–35.
Dias S, Wickramarachchi T, Sahabandu I, Escalante AA, Udagama PV. Population genetic structure of the Plasmodium vivax circumsporozoite protein (Pvcsp) in Sri Lanka. Gene. 2013;518:381–7.
Talha AA, Pirahmadi S, Mehrizi AA, Djadid ND, Nour BY, Zakeri S. Molecular genetic analysis of Plasmodium vivax isolates from Eastern and Central Sudan using pvcsp and pvmsp-3α genes as molecular markers. Infect Genet Evol. 2015;32:12–22.
Kaur H, Sehgal R, Goyal K, Makkar N, Yadav R, Bharti PK, et al. Genetic diversity of Plasmodium falciparum merozoite surface protein-1 (block 2), glutamate-rich protein and sexual stage antigen Pfs25 from Chandigarh. North India Trop Med Int Health. 2017;22:1590–8.
Simon B, Sow F, Al Mukhaini SK, Al-Abri S, Ali OA, Bonnot G, et al. An outbreak of locally acquired Plasmodium vivax malaria among migrant workers in Oman. Parasite. 2017;24:25.
Liu Y, Zhou RM, Zhang YL, Wang DQ, Li SH, Yang CY, Qian D, Zhao YL, Zhang HW, Xu BL. Analysis of polymorphisms in the circumsporozoite protein gene of Plasmodium vivax isolates from Henan Province. China Malar J. 2018;17:103.
Kaur H, Sehgal R, Kumar A, Sehgal A, Bharti PK, Bansal D, et al. Exploration of genetic diversity of Plasmodium vivax circumsporozoite protein (Pvcsp) and Plasmodium vivax sexual stage antigen (Pvs25) among North Indian isolates. Malar J. 2019;18:308.
Võ TC, Lê HG, Kang JM, Moe M, Naw H, Myint MK, et al. Genetic polymorphism and natural selection of circumsporozoite protein in Myanmar Plasmodium vivax. Malar J. 2020;19:303.
Alves RT, Póvoa MM, Goldman IF, Cavasini CE, Rossit AR, Machado RL. A new polymerase chain reaction/restriction fragment length polymorphism protocol for Plasmodium vivax circumsporozoite protein genotype (VK210, VK247, and P. vivax-like) determination. Diagn Microbiol Infect Dis. 2007;59:415–9.
Patil A, Orjuela-Sánchez P, da Silva-Nunes M, Ferreira MU. Evolutionary dynamics of the immunodominant repeats of the Plasmodium vivax malaria-vaccine candidate circumsporozoite protein (CSP). Infect Genet Evol. 2010;10:298–303.
Rosenberg R, Wirtz RA, Lanar DE, Sattabongkot J, Hall T, Waters AP, et al. Circumsporozoite protein heterogeneity in the human malaria parasite Plasmodium vivax. Science. 1989;245:973–6.
Leclerc MC, Durand P, Gauthier C, Patot S, Billotte N, Menegon M, et al. Meager genetic variability of the human malaria agent Plasmodium vivax. Proc Natl Acad Sci USA. 2004;101:14455–60.
Zakeri S, Mehrizi AA, Djadid ND, Sounou G. Circumsporozoite protein gene diversity among temperate and tropical Plasmodium vivax isolates from Iran. Trop Med Int Health. 2006;11:729–37.
Qari SH, Shi YP, Goldman IF, Udhaykumar V, Collins WE, Lal AA, et al. Identification of Plasmodium vivax-like human malaria parasite. Lancet. 1993;341:780–3.
Tahar R, Ringwald P, Basco LK. Heterogeneity in the circumsporozoite protein gene of Plasmodium malariae isolates from sub-Saharan Africa. Mol Biochem Parasitol. 1998;92:71–8.
Birkenmeyer L, Muerhoff AS, Dawson GJ, Desai SM. Isolation and characterization of the MSP1 genes from Plasmodium malariae and Plasmodium ovale. Am J Trop Med Hyg. 2010;82:996–1003.
Zakeri S, Raeisi A, Afsharpad M, Kakar Q, Ghasemi F, Atta H, et al. Molecular characterization of Plasmodium vivax clinical isolates in Pakistan and Iran using pvmsp-1, pvmsp-3α and Pvcsp genes as molecular markers. Parasitol Int. 2010;59:15–21.
Bonilla JA, Validum L, Cummings R, Palmer CJ. Genetic diversity of Plasmodium vivax pvcsp and pvmsp1 in Guyana, South America. Am J Trop Med Hyg. 2006;75:830–5.
Kibria MG, Elahi R, Mohon AN, Khan WA, Haque R, Alam MS. Genetic diversity of Plasmodium vivax in clinical isolates from Bangladesh. Malar J. 2015;14:267.
Imwong M, Pukrittayakamee S, Grüner AC, Rénia L, Letourneur F, Looareesuwan S, et al. Practical PCR genotyping protocols for Plasmodium vivax using pvcs and pvmsp1. Malar J. 2005;4:20.
Putaporntip C, Jongwutiwes S, Sakihama N, Ferreira MU, Kho WG, Kaneko A, et al. Mosaic organization and heterogeneity in frequency of allelic recombination of the Plasmodium vivax merozoite surface protein-1 locus. Proc Natl Acad Sci USA. 2002;99:16348–53.
Thanapongpichat S, Khammanee T, Sawangjaroen N, Buncherd H, Tun AW. Genetic diversity of Plasmodium vivax in clinical isolates from Southern Thailand using PvMSP1, PvMSP3 (PvMSP3α, PvMSP3β) genes and eight microsatellite markers. Korean J Parasitol. 2019;57:469.
Sheikh IM, Pasha MK, Williams VS, Raza SQ, Khan KS. Environmental geology of the Islamabad-Rawalpindi area, northern Pakistan. Regional Studies of the Potwar-Plateau Area, Northern Pakistan. USAID, Geological Survey of Pakistan. Bulletin 2078-G. 2008.
Qureshi NA, Fatima H, Afzal M, Khattak AA, Nawaz MA. Occurrence and seasonal variation of human Plasmodium infection in Punjab Province. Pakistan BMC Infect Dis. 2019;19:935.
Russell DW, Sambrook J. The condensed protocols from Molecular Cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; 2006.
Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2.
Kumar S, Stecher G, Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Khatoon L, Baliraine FN, Bonizzoni M, Malik SA, Yan G. Prevalence of antimalarial drug resistance mutations in Plasmodium vivax and P. falciparum from a malaria-endemic area of Pakistan. Am J Trop Med Hyg. 2009;81:525–8.
Khatoon L, Baliraine FN, Bonizzoni M, Malik SA, Yan G. Genetic structure of Plasmodium vivax and Plasmodium falciparum in the Bannu district of Pakistan. Malar J. 2010;9:112.
Ord R, Polley S, Tami A, Sutherland CJ. High sequence diversity and evidence of balancing selection in the Pvmsp3α gene of Plasmodium vivax in the Venezuelan Amazon. Mol Biochem Parasitol. 2005;144:86–93.
Shabani SH, Zakeri S, Mehrizi AA, Mortazavi Y, Djadid ND. Population genetics structure of Plasmodium vivax circumsporozoite protein during the elimination process in low and unstable malaria transmission areas, southeast of Iran. Acta Trop. 2016;160:23–34.
González-Cerón L, Martinez-Barnetche J, Montero-Solís C, Santillán F, Soto AM, Rodríguez MH, et al. Molecular epidemiology of Plasmodium vivax in Latin America: polymorphism and evolutionary relationships of the circumsporozoite gene. Malar J. 2013;12:243.
Moon SU, Lee HW, Kim JY, Na BK, Cho SH, Lin K, et al. High frequency of genetic diversity of Plasmodium vivaxfield isolates in Myanmar. Acta Trop. 2009;109:30–6.
Cui LW, Mascorro CN, Fan Q, Rzomp KA, Khuntirat B, Zhou G, et al. Genetic diversity and multiple infections of Plasmodium vivaxmalaria in western Thailand. Am J Trop Med Hyg. 2003;685:613–9.
Chenet SM, Tapia LL, Escalante AA, Durand S, Lucas C, Bacon DJ. Genetic diversity and population structure of genes encoding vaccine candidate antigens of Plasmodium vivax. Malar J. 2012;11:68.
González-Cerón L, Rodríguez MH, Nettel-Cruz JA, Hernández-Ávila JE, Malo-García IR, Santillán-Valenzuela F, et al. Plasmodium vivax CSP-Pvs25 variants from southern Mexico produce distinct patterns of infectivity for Anopheles albimanus versus An. pseudopunctipennis, in each case independent of geographical origin. Parasit Vectors. 2019;12:86.
Hughes AL. The evolution of amino acid repeat arrays in Plasmodium and other organisms. J Mol Evol. 2004;59:528–35.
Parobek CM, Bailey JA, Hathaway NJ, Socheat D, Rogers WO, Juliano JJ. Differing patterns of selection and geospatial genetic diversity within two leading Plasmodium vivax candidate vaccine antigens. PLoS Negl Trop Dis. 2014;8:e2796.
Lopez AC, Ortiz A, Coello J, Sosa-Ochoa W, Torres RE, Banegas EI, et al. Genetic diversity of Plasmodium vivax and Plasmodium falciparum in Honduras. Malar J. 2012;11:391.
Kim SH, Hwang SY, Shin JH, Moon CS, Kim DW, Kho WG. Molecular genetic characterization of the merozoite surface protein 1 Gene of Plasmodium vivax from reemerging Korean isolates. Clin Vaccine Immunol. 2009;16:733–8.
Putaporntip C, Hongsrimuang T, Seethamchai S, Kobasa T, Limkittikul K, Cui L, et al. Differential prevalence of Plasmodium infections and cryptic Plasmodium knowlesi malaria in humans in Thailand. J Infect Dis. 2009;199:1143–50.
Kim JR, Imwong M, Nandy A, Chotivanich K, Nontprasert A, Tonomsing N, et al. Genetic diversity of Plasmodium vivax in Kolkata. India Malar J. 2006;5:71.
Barry AE, Waltmann A, Koepfli C, Barnadas C, Mueller I. Uncovering the transmission dynamics of Plasmodium vivax using population genetics. Pathog Glob Health. 2015;109:142–52.
Acknowledgements
We would like to thank medical staff of hospital and all the study participants to participate in the study.
Funding
There was no financial support for this research.
Author information
Authors and Affiliations
Contributions
ZB conducted laboratory experiments and data analysis, AF helped in sample collection, clinical inputs and malaria identification, RR helped in primer designing and molecular data input, AM provided scientific inputs and critical review of manuscript, SK helped specifically in CSP gene analysis, SN supervised the wet labs, critically reviewed manuscript, helped in manuscript drafting and data analysis, SW conceived the idea and study design, provided all wet lab reagents/lab facilities and contributed to the critical revision of the manuscript,. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was approved by institutional review board (IRB), Virtual University of Pakistan, (letter No. VU/ASRB/131–7). Blood samples were collected from diagnosed malaria patients after informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bibi, Z., Fatima, A., Rani, R. et al. Genetic characterization of Plasmodium vivax isolates from Pakistan using circumsporozoite protein (pvcsp) and merozoite surface protein-1 (pvmsp-1) genes as genetic markers. Malar J 20, 112 (2021). https://doi.org/10.1186/s12936-021-03654-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-021-03654-w