Abstract
Background
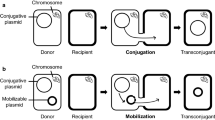
The conjugative plasmid, pLS20, isolated from Bacillus subtilis natto, has an outstanding capacity for rapid self-transfer. In addition, it can function as a helper plasmid, mediating the mobilization of an independently replicating co-resident plasmid.
Results
In this study, the oriT sequence of pLS20cat (oriTLS20) was eliminated to obtain the plasmid, pLS20catΔoriT. This resulted in the complete loss of the conjugative transfer of the plasmid but still allowed it to mobilize a co-resident mobilizable plasmid. Moreover, pLS20catΔoriT was able to mobilize longer DNA segments, up to 113 kb of chromosomal DNA containing oriTLS20, after mixing the liquid cultures of the donor and recipient for only 15 min.
Conclusions
The chromosomal DNA mobilization mediated by pLS20catΔoriT will allow us to develop a novel genetic tool for the rapid, easy, and repetitive mobilization of longer DNA segments into a recipient chromosome.
Similar content being viewed by others
Background
Bacterial cell division is asexual, and the genetic traits of mother and daughter cells with normal development are not changed fundamentally. However, horizontal gene transfer (HGT) can provide genetic plasticity for bacteria, even among different species [1]. HGT can lead sometimes to problematic effects for the recipient bacteria that accept the transferred genes. Those receiving undesirable genetic traits are excluded generally from the bacterial population, and it becomes less likely that harmful genes are inherited by the next generation. However, mobile genetic elements (MGE) capable of self-transfer and self-replication often confer genetic traits that are advantageous to the recipient bacterial cells, and that can be propagated rapidly within the population [2]. HGT includes at least three types of mechanisms, including transformation, transduction, and conjugation [1]. In the event of transformation, recipient cells uptake foreign DNAs depending on their own genetic competence, and the foreign DNAs do not necessarily have a MGE function. In transduction, foreign DNAs are transferred proactively together by bacteriophage infection, but the infection itself is often harmful for the recipient cell [3]. Conjugation refers to the transfer of DNA to a recipient bacterium from a donor bacterium through a mating event. During conjugation, the DNA transfers through dedicated conjugation machinery, encoded by genes on the conjugative DNA element. Conjugative DNA elements may be either plasmids or conjugative transposons, which are now more commonly referred to as integrative and conjugative elements or ICEs. HGT is environmentally important, as it increases the diversity of bacterial communities; however, it also serves as a useful tool for engineering desired bacterial strains via recombinant DNA technology for research and industrial purposes.
The typical mechanism for conjugative plasmid transfer between two bacterial strains proceeds as follows [4,5,6,7,8]. First, in the donor, a specific enzyme, relaxase, cleaves a phosphodiester bond of the plasmid DNA to produce a nick at a specific site on its “origin of transfer,” or oriT. As a result, relaxase is covalently linked to the 5′-end of the single-stranded plasmid DNA at the nick, and a DNA–protein complex, called relaxosome, is formed. Next, another specific protein, the type IV coupling protein (T4CP), interacts with the relaxosome and targets it for transfer through a type IV secretion system from the donor to the recipient. When one unit of the plasmid DNA is transferred completely into the recipient cell, the single-stranded DNA is circularized and begins to replicate as maturation of the double-stranded circular plasmid DNA occurs in the transconjugant.
Generally speaking, plasmids are classified into two types: conjugative and non-conjugative ones. The former can transfer itself to the recipient using self-coding specific enzymes and a secretion system that is necessary for the transfer to occur; the latter cannot transfer itself because it lacks the required set of genes to do so. However, non-conjugative plasmids containing the oriT sequence, or mobilizable plasmids, can be mobilized by a co-resident conjugative plasmid within the same cell, in a phenomenon known as mobilization. Mobilizable plasmids are believed to need the oriT and its cognate mob gene, the gene encoding relaxase [9, 10], but recently it was discovered that relaxase is not necessary for plasmid mobilization if a plasmid carries a “mimic” of the oriT region found in conjugative plasmids [11].
pLS20 is a 65-kbp-conjugative plasmid isolated from a strain of Bacillus subtilis natto [12]. It can transfer itself between various B. subtilis-related Gram positive bacteria, including Bacillus anthracis, Bacillus cereus, Bacillus licheniformis, Bacillus megaterium, Bacillus pumilus, and Bacillus thuringiensis [13]. pLS20cat, a derivative of pLS20 carrying a chloramphenicol resistance gene [14,15,16,17], possesses the incredible quality of being able to very rapidly transfer itself between cells within 15 min by simply mixing the liquid cultures containing donor and recipient cells [18]. In addition, pLS20cat can function as a helper plasmid, with the ability to mobilize an independently replicating and co-resident mobilizable plasmid containing a short oriT sequence from pLS20cat (oriTLS20) that is unaccompanied by its cognate mob gene [19].
In this study, we inactivated the oriTLS20 region of pLS20cat to obtain pLS20catΔoriT, rendering it completely immobile, but it was still able to facilitate mobilization of the plasmid containing the oriTLS20. Moreover, pLS20catΔoriT was able to mobilize longer chromosomal DNA segments containing oriTLS20 independently of the natural competence of the recipient cell. The larger DNA mobilization was achieved after mixing the liquid cultures of the donor and recipient for only 15 min. This pLS20catΔoriT-mediated chromosomal DNA mobilization will allow us to develop a novel genetic tool for rapid and repetitive accumulation of longer DNA segments into the recipient chromosome.
Methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Synthetic oligonucleotides used as PCR primers are shown in Table 2. Bacterial strains were grown on Lysogeny Broth (LB) medium (Difco) at 37 °C. When necessary, the medium was supplemented with antibiotics: 5 μg ml−1 chloramphenicol, 1 μg ml−1 erythromycin, 100 μg ml−1 spectinomycin, and 10 μg ml−1 kanamycin.
Construction of the recipient strain
The comK gene of strain 168 was inactivated by replacement with a spectinomycin resistance gene, as follows. Two DNA fragments, each of which corresponded to upstream and downstream regions of comK, were amplified by PCR using 168 DNA as a template with primers comK-uF/comK-uR for the upstream fragment and comK-dF/comK-dR for the downstream fragment (Table 2). Another DNA fragment containing the spectinomycin resistance gene of strain TMO310 (Table 1) was amplified using primers spc-F/spc-R (Table 2). The three fragments were ligated together by recombinant PCR using primers comK-uF/comK-dR to sandwich the spectinomycin resistance gene between the upstream and downstream regions of comK. The recombinant PCR fragment was transformed into strain 168 conferring spectinomycin resistance and yielding the new strain, YNB001 (comK::spc), which was used as the recipient for the conjugative DNA transfer in this study.
Construction of pLS20catΔoriT
The oriTLS20 region of pLS20cat was inactivated by marker-free deletion, as previously described [20]. Two DNA fragments corresponding to the upstream (fragment 1) and downstream (fragment 2) regions of oriTLS20 were amplified by PCR using pLS20cat DNA as the template with primers oriT-uF/oriT-uR for the upstream region, and oriT-dF/oriT-dR for the downstream region (Table 2). Because the tail of fragment 1 and the head of fragment 2 were identical for 30 bp, those regions were responsible for the later deletion of the oriTLS20 region by intramolecular recombination. Another DNA fragment of the mazF kan cassette (fragment 3) was amplified from TMO311 DNA (Table 1) using primers mazF-F/mazF-R (Table 2). The three PCR fragments were designed to be connected in the order 1–3–2 by recombinant PCR using primers oriT-uF/oriT-dR. The recombinant PCR fragment was transformed then into strain PKS11 (Table 1) to confer kanamycin resistance, obtaining the new strain YNB022, in which pLS20cat was altered by integrating the PCR fragment through a double crossover event at the oriTLS20 region. YNB022 was grown overnight at 37 °C in LB liquid medium containing kanamycin. An aliquot of the culture was transferred into a fresh LB liquid medium containing 1 mM isopropyl-thiogalactopyranoside (IPTG) and the cells were allowed to grow for 2 h at 37 °C. Then, an aliquot of the culture was spread on an LB plate containing 1 mM IPTG and incubated at 37 °C overnight. In the presence of IPTG, mazF was expressed, producing a suicidal toxin so that only the cells that could pop-out the mazF kan cassette through intramolecular recombination could survive. Of those colonies appearing on the plate, kanamycin-sensitive colonies were sequenced to confirm the correct deletion of the oriTLS20 region. The resulting plasmid was designated as pLS20catΔoriT.
Construction of the donor strains
The donor strain YNB060 was constructed as follows (Fig. 1). Two fragments corresponding to upstream (fragment 1) and downstream (fragment 4) regions of yhfM were amplified from 168 DNA using primers yhfM-uF/yhfM-uR1 (for upstream) and yhfM-dF/yhfM-dR (for downstream) (Table 2). Fragment 2 containing the oriTLS20 was amplified using pLS20cat as a template with primers oriT-F/oriT-R (Table 2). In addition, fragment 3 carrying the erythromycin resistance gene was amplified using plasmid pMutin2 as the template with primers erm-F1/erm-R (Table 2). Fragments 1–4 were ligated in the order 1–2–3–4 by recombinant PCR using primers yhfM-uF/yhfM-dR. Strain TMO311 (aprE::kan) was transformed with the recombinant PCR fragment to select colonies resistant both to erythromycin and kanamycin. The resulting strain was designated as YNB060 (Table 1), which had the erythromycin marker with oriTLS20 and kanamycin marker at both the yhfM and aprE loci, located 6.6 kb apart from each other on the same chromosome. In addition, the direction of replication of oriTLS20 was oriented toward the kanamycin marker located 6.6 kb downstream.
Schematic representation of the integration of the loci of oriTLS20 and the kanamycin resistance gene of the donor strains. The gene loci where oriTLS20 was integrated and the distances from the aprE loci where the kanamycin resistance gene (kan) was integrated are shown (top). The oriTLS20 regions in donor strains are shown with forward- (oriT-F) and reverse- (oriT-R) oriented arrowheads (middle). Names of strains are aligned underneath the corresponding integration loci of oriTLS20 (bottom)
Another strain, YNB061, was constructed similarly as described above. Two fragments of yhfM, representing upstream and downstream regions, were amplified using primers yhfM-uF/yhfM-uR2 and yhfM-dF/yhfM-dR (Table 2), respectively. The oriTLS20 fragment (fragment 2) and the erythromycin resistance fragment (fragment 3) were amplified using primers oriT-F/oriT-R and erm-F2/erm-R (Table 2), respectively. The four fragments were ligated by recombinant PCR using primers yhfM-uF/yhfM-dR. Strain TMO311 was transformed with the recombinant PCR fragment selecting erythromycin-resistant colonies as YNB061 (Table 1). In contrast to YNB060, in YNB061 the direction of replication of oriTLS20 on the yhfM locus was oriented oppositely to the kanamycin marker on the aprE locus.
The other additional strains, YNB069, YNB062, and YNB097, were constructed similarly to those described above (Fig. 1). For YNB069, two fragments of upstream (fragment 1) and downstream (fragment 4) regions of yhfK were amplified from 168 DNA using primers yhfK-uF/yhfK-uR1 and yhfK-dF/yhfK-dR (Table 2), respectively. For YNB062, two fragments of upstream and downstream regions of yhfC were amplified using primers yhfC-uF/yhfC-uR1 and yhfC-dF/yhfC-dR (Table 2), respectively. For YNB097, two fragments of upstream and downstream regions of yhcT were amplified using primers yhcT-uF/yhcT-uR1 and yhcT-dF/yhcT-dR (Table 2), respectively. The oriTLS20 fragment (fragment 2) and the erythromycin resistance fragment (fragment 3) were the same as those used above for YNB060 construction. For each case, the respective four fragments were ligated by recombinant PCR using primers yhfK-uF/yhfK-dR for YNB069, primers yhfC-uF/yhfC-dR for YNB062, and primers yhcT-uF/yhcT-dR for YNB097 (Table 2). Each of the recombinant PCR fragments was used to transform TMO311 (aprE::kan) to select colonies resistant to both erythromycin and kanamycin. The resulting strains were designated as YNB069, YNB062, and YNB097 (Table 1), which all had the erythromycin marker with oriTLS20 at the yhfK, yhfC, and yhcT loci, and the kanamycin marker at the aprE locus set apart from each other by 9.5, 16.4, and 113 kb within the chromosome, respectively. In addition, in all these strains, the direction of replication of oriTLS20 was forward-oriented to the kanamycin marker.
Conjugative DNA mobilization
Conjugative DNA mobilization was performed in the liquid medium, as previously described [18]. Donor and recipient strains were cultured independently overnight in 5 ml of LB liquid medium containing the appropriate antibiotics at 37 °C with shaking at 180 rpm. Each of the cultures was diluted to an optical density for the cell of 0.05 at 600 nm (OD600) in 5 ml of fresh LB medium without antibiotics and incubated at 37 °C with shaking at 180 rpm. When OD600 reached 0.5–0.7, 500 μl of the donor and recipient cultures were mixed in a 1.5 ml microtube to stand at 37 °C for 15 min. The mixture was serially diluted and spread on LB plates containing various combinations of antibiotics to grow colonies overnight. On their respective plates, colonies were counted as colony forming units (CFU) of transformed recipients produced by conjugative transfer and mobilization (transconjugants) to calculate mobilization efficiencies [CFU of transconjugants/CFU of total recipients × 106 (ppm)].
Results
pLS20catΔoriT cannot transfer itself but can help to mobilize a co-resident plasmid carrying oriT LS20
As previously described [14,15,16], pLS20cat has the complete set of genes required for its own conjugative transfer. In fact, pLS20cat transfers itself from the donor PKS11 (Table 1, 168 with pLS20cat) or GR138 (strain 168 with pLS20cat and pGR16B) to the recipient YNB001 (comK::spc) within only 15 min after mixing the two parental liquid cultures, resulting in a large number (more than 2500 ppm) of recipient cells with acquired chloramphenicol resistance appearing as transconjugants (Fig. 2). It is also known that pLS20cat is capable of mobilizing a co-resident plasmid, pGR16B, carrying oriTLS20 and erythromycin resistance gene [19], as we observed the donor, GR138, confer erythromycin resistance on nearly 1000 ppm of recipient cells (Fig. 2). These results imply that the helper pLS20cat could be nearly twice more efficient at transforming recipient cells than the mobilizable plasmid pGR16B. In addition, about 100 ppm of the recipients obtained resistance to both erythromycin and chloramphenicol (Fig. 2), suggesting that about 10% of the transconjugants that accepted pGR16B also may have acquired pLS20cat.
Mobilization efficiencies of the mobilizable plasmid, pGR16B, and the helper plasmids, pLS20cat and pLS20catΔoriT. Liquid cultures of the recipient strain YNB001 (comK::spc) and one of the donor strains: PKS11 (168 with pLS20cat), YNB026 (168 with pLS20catΔoriT), GR138 (168 with pLS20cat and pGR16B), and YNB031 (168 with pLS20catΔoriT and pGR16B), were mixed for conjugative transfer and spread on LB plates containing both chloramphenicol and spectinomycin (CS), both erythromycin and spectinomycin (ES), chloramphenicol, erythromycin, and spectinomycin (CES), and spectinomycin alone. Colonies were counted as CFUs to calculate mobilization efficiencies [CFU of transconjugants (colonies on the CS, ES, and CES plates)/CFU of total recipients (colonies on the spectinomycin plate) × 106 (ppm)]. Values are means with standard deviations from three independent experiments. ND not detected (< 0.01 ppm). NP not performed
Since the bacterial cells carrying pLS20cat do not accept the pLS20cat-mediated genetic transfer [14,15,16], the transconjugants that have accepted pLS20cat could not be transformed again using the same conjugative transfer system. On the other hand, the cells carrying pLS20cat could transfer not only pLS20cat itself but also mobilize co-resident pGR16B to other strains further. If these transconjugants were released into the environment, the antibiotic resistance genes would be spread to other bacterial cells, causing the undesirable emergence of new antibiotic-resistant bacteria [21, 22]. To avoid the self-transfer of pLS20cat, we aimed at knocking-out oriTLS20 in pLS20cat to construct pLS20catΔoriT. As expected, the donor YNB026 did not transfer pLS20catΔoriT at all, whereas YNB031 mobilized the co-resident pGR16B to confer erythromycin resistance on the recipients (Fig. 2). Furthermore, the mobilization efficiency of pGR16B was nearly the same whether pLS20cat or pLS20catΔoriT served as the helper plasmid. These results indicate that the pLS20cat-dependent mobilization of pGR16B did not require self-mobility of the helper plasmid, pLS20cat. In addition, knocking-out oriTLS20 in pLS20cat did not affect the mobilization efficiency of the co-resident, pGR16B.
pLS20catΔoriT can mobilize chromosomal DNA containing oriT LS20
As shown above and in previous studies [19], pLS20cat can efficiently mobilize the co-resident mobilizable plasmid with oriTLS20 but without its cognate mob gene. Recently, conjugative transfer was shown to mobilize a large DNA fragment, representing the entire chromosome of Mycoplasma [23]. Thus, we conceived the idea that pLS20cat may be able to mobilize chromosomal DNA, depending on the status of the oriTLS20 region.
In the donor chromosome, oriTLS20 was introduced at the yhfM locus, 6.6 kb upstream of the kanamycin resistance gene at the aprE locus; in strains YNB060 and YNB061 (Table 1), the direction of replication of oriTLS20 was forward- and reverse-oriented to the kanamycin resistance gene, respectively. pLS20cat or pLS20catΔoriT was introduced into the donor as the helper plasmid, yielding these new strains: (1) YNB065 (YNB060 with pLS20cat), (2) YNB066 (YNB061 with pLS20cat), (3) YNB091 (YNB060 with pLS20catΔoriT), and (4) YNB095 (YNB061 with pLS20catΔoriT). On the other hand, in the recipient strain YNB001, comK encoding the key transcription factor for natural competence was inactivated so that the strain completely lost its natural competence (data not shown).
Strains YNB065 and YNB066, both carrying pLS20cat, conferred chloramphenicol resistance on more than 2300 ppm of the recipients (Fig. 3), but YNB065 was able to confer kanamycin resistance on only 1 ppm of the recipient cells; YNB066 did not confer kanamycin resistance at all (Fig. 3). These results indicate that pLS20cat could transfer the kanamycin resistance gene located 6.6 kb downstream of oriTLS20, if the direction of oriTLS20 replication was forward-oriented to the kanamycin resistance gene. In addition, since the recipients had no natural competence, the acquisition of kanamycin resistance depended solely on the conjugative transfer. On the other hand, YNB065 conferred not only kanamycin resistance but also chloramphenicol resistance, on nearly 1 ppm of the recipients (Fig. 3). These results suggest that a large majority of the kanamycin-resistant recipients that accepted the chromosomal DNA could additionally have acquired the helper plasmid, pLS20cat.
Mobilization efficiencies of the kanamycin resistance gene at the aprE locus and the helper plasmids, pLS20cat and pLS20catΔoriT. YNB060 and YNB061 have oriTLS20 forward- and reverse-oriented to the kanamycin resistance gene (6.6F and R, respectively). Liquid cultures of the recipient strain YNB001 (comK::spc) and one of the donor strains: YNB065 (YNB060 with pLS20cat), YNB091 (YNB060 with pLS20catΔoriT), YNB066 (YNB061 with pLS20cat), YNB095 (YNB061 with pLS20catΔoriT), were mixed for conjugative transfer and spread on LB plates containing both chloramphenicol and spectinomycin (CS), both kanamycin and spectinomycin (KS), chloramphenicol, kanamycin, and spectinomycin (CKS), and spectinomycin alone. Colonies were counted as CFUs to calculate mobilization efficiencies [CFU of transconjugants (colonies on the CS, KS, and CKS plates)/CFU of total recipients (colonies on the spectinomycin plate) × 106 (ppm)]. Values are means with standard deviations from three independent experiments. ND not detected (< 0.01 ppm)
When pLS20catΔoriT was introduced as the helper plasmid into YNB060 (YNB091) and YNB061 (YNB095), it appeared that no recipient acquired chloramphenicol resistance (Fig. 3), confirming the loss of self-mobility in pLS20catΔoriT. On the other hand, and more importantly, when YNB091 was used as the donor, pLS20catΔoriT transferred kanamycin resistance to recipients with a similar efficiency and oriTLS20-orientation dependency to pLS20cat (Fig. 3). These results clearly indicate that pLS20catΔoriT can exert its helper activity as efficiently as the original helper plasmid, pLS20cat, not only in the case of mobilizing pGR16B but also when mobilizing the chromosomal DNA. YNB095 did not confer kanamycin resistance on the recipients, confirming that the DNA mobilization depended on the forward-oriented oriTLS20.
The distance between oriTLS20 and the kanamycin marker was extended by 9.5 kb, 16.4 kb, and 113 kb in a stepwise fashion in strains YNB069, YNB062, and YNB097, respectively (Fig. 1). In all of these strains, the direction of replication of oriTLS20 was forward-oriented to the kanamycin resistance gene. A helper plasmid, either pLS20cat or pLS20catΔoriT, was introduced into each donor to create new donor strains: YNB071 (YNB069 with pLS20cat), YNB067 (YNB062 with pLS20cat), YNB099 (YNB097 with pLS20cat), YNB092 (YNB069 with pLS20catΔoriT), YNB094 (YNB062 with pLS20catΔoriT), and YNB100 (YNB097 with pLS20catΔoriT). All donors with pLS20cat conferred chloramphenicol resistance on more than 600 ppm of recipient cells, whereas the other donors, with pLS20catΔoriT, did not confer chloramphenicol resistance at all (Fig. 4). However, and more importantly, all of the strains with pLS20catΔoriT were able to confer kanamycin resistance on 0.5–10.0 ppm of the recipient cells. Efficiencies were nearly equivalent to those achieved with YNB091 as the donor (Fig. 4), indicating that the length of mobilized DNA could be extended at least to 113 kb. In addition, pLS20catΔoriT exhibited similar efficiencies to pLS20cat when mobilizing longer segments of chromosomal DNA (Fig. 4). These results also indicate that self-mobility of pLS20cat was not necessary for its helper function for mobilizing longer segments of chromosomal DNA.
Mobilization efficiencies of the kanamycin resistance gene at the aprE locus and the helper plasmids, pLS20cat and pLS20catΔoriT. Liquid cultures of the recipient strain YNB001 (comK::spc) and one of the donor strains: YNB065 (YNB060 with pLS20cat), YNB091 (YNB060 with pLS20catΔoriT), YNB071 (YNB069 with pLS20cat), YNB092 (YNB069 with pLS20catΔoriT), YNB067 (YNB062 with pLS20cat), YNB094 (YNB062 with pLS20catΔoriT), YNB099 (YNB097 with pLS20cat), and YNB100 (YNB097 with pLS20catΔoriT), were mixed for conjugative transfer of long DNA segments (6.6F, 9.5F, 16.4F, and 113F for 6.6, 9.5, 16.4, and 113 kb between oriTLS20 and the kanamycin marker, respectively), and spread on LB plates containing both chloramphenicol and spectinomycin (CS), both kanamycin and spectinomycin (KS), and spectinomycin alone. Colonies were counted as CFUs to calculate mobilization efficiencies [CFU of transconjugants (colonies on the CS and KS plates)/CFU of total recipients (colonies on the spectinomycin plate) × 106 (ppm)]. Values are means with standard deviations from three independent experiments. ND not detected (< 0.01 ppm)
Discussion
Strains of B. subtilis 168 derivatives have an advantage over other bacteria because their natural competence and high recombination efficiency allow for plasticity of their genome. A number of artificial introductions and compilations of various sizes and kinds of DNA segments have been performed successfully into the B. subtilis genome [24]. Therefore, B. subtilis has been regarded, generally, as one of the most promising platforms for designing, assembling, and modifying synthetic DNA, or even on larger scales with an entire synthetic genome. Accordingly, there is increasing demand of novel genetic tools for mobilizing longer DNA segments, which will push forward the research and development in synthetic genome approaches [25].
Here we demonstrated that pLS20cat conjugative transfer was capable of mobilization of not only a mobilizable plasmid carrying oriTLS20 but also chromosomal DNA. In this study, however, both the donors and recipients were derived from the same parental strain, B. subtilis 168, and recombination events between the chromosome and the mobilized DNA could occur at any homologous locations; therefore, we are not able to state that the entire length of mobilized DNA accurately replaced the respective part of the chromosome. Nevertheless, we can assume, at least, that the mobilization of chromosomal DNA initiated at the integration point of oriTLS20 and continued until the kanamycin marker because mobilization was seen only when the direction of replication of oriTLS20 was forward-oriented to the kanamycin resistance gene (Fig. 3). Furthermore, the present results indicate that DNA segments up to 113 kb could be mobilized (Fig. 4), which may be one of the longest segments of DNA mobilized artificially from one cell to another within such a short period of only 15 min. For further applications, it would be worthwhile to test various conditions in order to extend the length of mobilizable DNA.
As mentioned above, the mobilization of chromosomal DNA depended upon the forward-orientation of the oriTLS20 replication region, implying that the replication origin may function unidirectionally; however, a previous study suggested the possibility that oriTLS20 may be able to replicate bidirectionally [19]. If this is true, it is likely that oriTLS20 replication in the reverse direction would be too weak to enable the mobilization of longer DNA segments. On the other hand, the reverse-oriented oriTLS20 could lead the counterclockwise replication of the other side of chromosome, which requires synthesis of much longer DNA to encounter the kanamycin marker. As we failed to detect any kanamycin-resistant transformant using the reverse-oriented oriTLS20, there could be a certain limit in the length of mobilizable DNA, which is yet to be defined.
We inactivated the oriTLS20 of pLS20cat to make pLS20catΔoriT, which never transferred itself between cells but was able to mobilize longer segments of chromosomal DNA with nearly the same efficiency as the self-transfer of pLS20cat. To our knowledge, this is the first demonstration that the oriT function of pLS20cat is not required for performing its helper function in mobilizing DNA fragments containing oriTLS20. The DNA mobilization by the pLS20cat conjugative system only occurs to a recipient not harboring pLS20cat [14,15,16]. As described above, when donors with pLS20cat were used, more than 2000 ppm of the recipient cells became chloramphenicol resistant by accepting pLS20cat. On the other hand, the transfer of kanamycin resistance was seen for only 1–10 ppm of recipients. These results imply that nearly all the recipients that acquired the chromosomal DNA also could have accepted pLS20cat. Therefore, recipients that previously acquired chromosomal DNA with the help of pLS20cat could no longer accept a conjugative DNA mobilization based on the pLS20 system. On the other hand, recipients that acquired chromosomal DNA with the help of pLS20catΔoriT did not have pLS20catΔoriT and could accept new rounds of DNA mobilization. This acquired knowledge will be beneficial for developing a novel genetic tool for repetitive accumulation of longer DNA segments into the recipient chromosome. Furthermore, the pLS20catΔoriT also would be useful for transforming other B. subtilis-related Gram positive bacteria, including B. anthracis, B. cereus, B. licheniformis, B. megaterium, B. pumilus, and B. thuringiensis [13].
Conclusions
In this study, the oriTLS20 region of pLS20cat was eliminated to obtain pLS20catΔoriT, which resulted in completely eliminating the plasmid’s own mobility, while maintaining an ability to efficiently mediate the conjugative mobilization of a neighboring mobilizable plasmid. Moreover, pLS20catΔoriT was able to mobilize longer DNA segments, up to 113 kb of chromosomal DNA, containing the oriTLS20 region after mixing the liquid cultures of the donor and recipient for only 15 min. Understanding this chromosomal DNA mobilization by pLS20catΔoriT will allow us to develop a novel genetic tool for the rapid, easy, and repetitive accumulation of longer DNA segments into a recipient chromosome.
Abbreviations
- CFU:
-
colony forming units
- HGT:
-
horizontal gene transfer
- IPTG:
-
isopropyl-thiogalactopyranoside
- LB:
-
Lysogeny Broth
- MGE:
-
mobile genetic elements
- T4CP:
-
type IV coupling protein
- PCR:
-
polymerase chain reaction
References
Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–32.
Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–21.
Davison J. Genetic exchange between bacteria in the environment. Plasmid. 1999;42:73–91.
Abajy MY, Kopeć J, Schiwon K, Burzynski M, Döring M, Bohn C, et al. A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in Gram-positive bacteria. J Bacteriol. 2007;189:2487–96.
Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808.
Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:277–301.
Wallden K, Rivera-Calzada A, Waksman G. Type IV secretion systems: versatility and diversity in function. Cell Microbiol. 2010;12:1203–12.
Zechner EL, Lang S, Schildbach JF. Assembly and mechanisms of bacterial type IV secretion machines. Philos Trans R Soc B. 2012;367:1073–87.
Francia MV, Varsaki A, Garcillán-Barcia MP, Latorre A, Drainas C, De La Cruz F. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 2004;28:79–100.
Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EPC, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74:434–52.
Ramsay JP, Firth N. Diverse mobilization strategies facilitate transfer of non-conjugative mobile genetic elements. Curr Opin Microbiol. 2017;38:1–9.
Tanaka T, Kuroda M, Sakaguchi K. Isolation and characterization of four plasmids from Bacillus subtilis. J Bacteriol. 1977;129:1487–94.
Koehler TM, Thorne CB. Bacillus subtilis (natto) plasmid pLS20 mediates interspecies plasmid transfer. J Bacteriol. 1987;169:5271–8.
Meijer WJJ, Boer AJ, Tongeren SV, Venema G, Bron S. Characterization of the replication region of the Bacillus subtilis plasmid pLS20: a novel type of replicon. Nucleic Acids Res. 1995;23:3214–23.
Singh PK, Ramachandran G, Durán-Alcalde L, Alonso C, Wu LJ, Meijer WJJ. Inhibition of Bacillus subtilis natural competence by a native, conjugative plasmid-encoded comK repressor protein. Environ Microbiol. 2012;14:2812–25.
Singh PK, Ramachandran G, Ramos-Ruiz R, Peiro-Pastor R, Abia D, Wu LJ, et al. Mobility of the native Bacillus subtilis conjugative plasmid pLS20 is regulated by intercellular signaling. PLoS Genet. 2013;9:10.
Ramachandran G, Singh PK, Luque-Ortega JR, Yuste L, Alfonso C, Rojo F, Wu LJ, Meijer WJJ. A complex genetic switch involving overlapping divergent promoters and DNA looping regulates expression of conjugation genes of a Gram-positive plasmid. PLoS Genet. 2014;10:10.
Itaya M, Sakaya N, Matsunaga S, Fujita K, Kaneko S. Conjugational transfer kinetics of pLS20 between Bacillus subtilis in liquid medium. Biosci Biotechnol Biochem. 2006;70:740–2.
Ramachandran G, Miguel-Arribas A, Abia D, Singh PK, Crespo I, Gago-Córdoba C, et al. Discovery of a new family of relaxases in Firmicutes bacteria. PLoS Genet. 2017;13:2.
Morimoto T, Ara K, Ozaki K, Ogasawara N. A new simple method to introduce marker-free deletions in the Bacillus subtilis genome. Genes Genet Syst. 2009;84:315–8.
Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–33.
Berglund B. Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. Infect Ecol Epidemiol. 2015;5:28564.
Dordet-Frisoni E, Sagné E, Baranowski E, Breton M, Nouvel LX, Blanchard A, et al. Chromosomal transfers in mycoplasmas: when minimal genomes go mobile. MBio. 2014;5:6.
Itaya M, Fujita K, Kuroki A, Tsuge K. Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat Methods. 2008;5:41–3.
Richardson SM, Mitchell LA, Stracquadanio G, Yang K, Dymond JS, DiCarlo JE, et al. Design of a synthetic yeast genome. Science. 2017;355:1040–4.
Authors’ contributions
KY and WM conceived the idea for the project and initiated the study in CSIC-UAM with the MEXT grant of Open Partnership Joint Research Projects/Seminars. MM conducted most of the experiments and analyzed the results, under the supervision and with indispensable assistance of KT in strain construction, SI in data analysis, and ST in conjugation experiments in Kobe, and with indispensable assistance of AA in plasmid preparation in Madrid. KY wrote the final manuscript with MM. All authors read and approved the final manuscript.
Acknowledgements
The authors thank Mr. Jorge Val Calvo for his help in experiments.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets and materials obtained and analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan; in part by Special Coordination Funds for Promoting Science and Technology, Creation of Innovative Centers for Advanced Interdisciplinary Research Areas, and Open Partnership Joint Research Projects/Seminars. Work in the Meijer lab was funded by grants Bio2013-41489-P and BIO2016-77883-C2-1-P of the Ministry of Economy and Competitiveness of the Spanish Government to WM.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Miyano, M., Tanaka, K., Ishikawa, S. et al. Rapid conjugative mobilization of a 100 kb segment of Bacillus subtilis chromosomal DNA is mediated by a helper plasmid with no ability for self-transfer. Microb Cell Fact 17, 13 (2018). https://doi.org/10.1186/s12934-017-0855-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-017-0855-x