Abstract
Background
Currently, the structural characteristics of the swine major histocompatibility complex (MHC) class I molecule, also named swine leukocyte antigen class I (SLA-I) molecule need to be further clarified.
Results
A complex of SLA-I constituted by an SLA-2*HB01 molecule with swine β2-microglobulin and a cytotoxic T lymphocyte (CTL) epitope FMDV-AS64 (ALLRSATYY) derived from VP1 protein (residues 64–72) of Asia 1 serotype of foot-and-mouth disease virus (FMDV) was expressed, refolded, purified and crystallized. By preliminary X-ray diffraction analysis, it was shown that the diffraction resolution of the crystal was 2.4 Å and the space group belonged to P212121 with unit cell parameters a = 48.37, b = 97.75, c = 166.163 Å.
Conclusion
This research will be in favor of illuminating the structural characteristics of an SLA-2 molecule associated with a CTL epitope derived from Asia1 serotype of FMDV.
Similar content being viewed by others
Background
Class I of major histocompatibility complex (MHC) molecules are membrane-surface proteins, which are mainly responsible for binding and presenting endogenous antigenic peptides degraded from proteins coded by virus genome in target cells [1] or autoantigen [2]. These endogenous antigenic peptides usually constituted by 8–10 residues in length are derived from endogenous antigens degraded by cellular proteasome [3, 4]. When a peptide is complexed and correctly refolded with MHC class I molecule in the antigen presenting cells (APCs), it will be presented to the membrane surface of the APCs to intrigue the cytotoxic T lymphocytes (CTLs). On this occasion, the peptide is defined as a CTL epitope [5]. Once CTLs are activated by the CTL epitopes, CTLs will recognize the epitopes and then kill the infected cells [6]. Therefore, the expressed MHC class I molecules on membrane of APCs should contain three components: a polymorphic heavy chain (α chain) of class I, a monomorphic light chain of β2-microglobulin (β2m) noncovalently linking the α3 domain of the heavy chain and an epitope bound in a groove formed by the α1 and α2 domains of the heavy chain [7]. Analysis of structural characteristics of MHC class I is necessary so that the mechanism of antigen presentation associated with MHC class I molecules will be explored.
MHC class I genes in pigs (Sus scrofa domestica) located in the 7p1.1 band of the short arm of chromosome 7 are also named as swine leukocyte antigen class I (SLA-I) [8]. There are three constitutively expressed classical and polymorphic SLA-I genes in the genome, namely SLA-1, SLA-2 and SLA-3 [9]. Among them, SLA-2 is different from SLA-1 and SLA-3 in the N-terminal of their coding regions as previously described [10]. The swine β2m (sβ2m) is monomorphic and noncovalently links with the heavy chain of the SLA-I molecules, which bind a viral or an auto CTL epitope. Foot-and-mouth disease virus (FMDV) is a great danger to cloven-hoofed animals including pigs, because it can cause animals to develop an acute, febrile, and highly contagious infectious disease [11]. In FMDV, there are seven serotypes named as A, O, C, Asia1, SAT1, SAT2, and SAT3. However, none of them have mutual cross-immunity [12, 13]. Among them, the Asia1 serotype often occurs in Asian countries as previously reported [14, 15]. Therefore, to further epitope vaccine development, more CTL epitopes and their interactions with SLA-I should be investigated. Recently, crystal data of the SLA-1, SLA-2 and SLA-3 had been announced, and a few CTL epitopes derived from swine-origin influenza virus, O serotype of FMDV, Ebola virus and respiratory syndrome virus (PRRSV) were also discovered [16,17,18,19]. However, crystal of SLA-2 associated with CTL epitope derived from Asia1 serotype of FMDV remains elusive.
In this article, we introduce the expression, refolding, purification, crystallization and preliminary X-ray diffraction analysis of SLA-2*HB01 with an AS64 CTL epitope derived from the Aisa1 serotype of FMDV.
Methods
Expression and isolation of the proteins of SLA-2*HB01 and sβ2m
To construct the expression system of the SLA-2 haplotype HB01 allele (GenBank accession No. AB602431) coding for 275 amino acids in extracellular domain, a pair of primers was designed as shown in Table 1. The PCR product was recovered and cloned into pMD®19-T simple vector as previously described [10, 19]. After identification by digestion with Nde I and Xho I and followed by sequencing, the interest of SLA-2*HB01 was further cloned into the pET21a (+) vector. The recombinant pET21a (+) containing the sβ2m had been done in our laboratory previously [19]. The two recombinant SLA-2*HB01 and sβ2m plasmids were induced to express in BL21 (Rosetta) Escherichia coli strain. The inclusion bodies were extracted as follows [17, 19]: In 2 L Luria–Bertani medium (LB), the SLA-2*HB01 and sβ2m expression strains were inoculated and incubated at 180 rpm for 3–4 h in a shaking incubator at 37 °C until the OD600 value reached 0.5–0.6. Then, a final concentration of 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was used in the medium to induce the interest of proteins to express in same cultivating environment as above. After 5 h, the bacteria were collected and cooled at 4 °C for 30 min. Then the bacteria were centrifugated for 15 min at 6000 rpm at 4 °C to collect pellets followed by washing them for three times with a solution buffer consisting of 50 mM Tris–HCl, 100 mM NaCl, 10 mM EDTA, 0.5% (v/v) Triton X-100, 1 mM DTT. In the end, a guanidinium chloride buffer [6 M guanidinium chloride, 100 mM NaCl, 10 mM EDTA, 50 mM Tris–HCl pH 8.0, 10% (v/v) glycerine, 10 mM DTT] was used to dissolve the isolated inclusion body proteins to a final concentration of 30 mg/mL.
Refolding and purification of the SLA-2*HB01-AS64-sβ2m complex
The refolded SLA-I (SLA-2*HB01-AS64-sβ2m) complex was carried out as described previously [6] with improvement recommended by Feng et al. [19]. Firstly, the epitope AS64 (ALLRSATYY) derived from the VP1 protein residues 64–72 of Asia1 FMDV was firstly dissolved in dimethyl sulfoxide (DMSO) and then diluted in water. The inclusion body proteins of SLA-2*HB01 and sβ2m were renatured and refolded with the AS64 epitope according to a 1:1:3 molar ratio by using the gradual dilution method in a refolding buffer (100 mM Tris pH 8.0, 400 mM L-Arg HCl, 2 mM EDTA, 5 mM GSH, 0.5 mM GSSH, 0.5 mM PMSF) at 4 °C. After refolding for 24 h, the soluble SLA-2*HB01-AS64-sβ2m complex was further concentrated and purified by chromatography separation on a Superdex 200 16/60 HiLoad size-exclusion column (GE Healthcare) followed by an anion-exchange chromatography Resource Q (GE Healthcare). SLA-2*HB01-AS64-sβ2m complex information is shown in Table 1.
Crystallizing the SLA-2*HB01-AS64-sβ2m complex
The final purified SLA-2*HB01-AS64-sβ2m complex proteins (~ 44 kDa) were further dialyzed against a crystallizing buffer (50 mM NaCl, 20 mM Tris–HCl pH 8.0) overnight at 4 °C and then concentrated to 15 mg/mL. Crystallizing assays were tried with Index and Crystal Screen kit (Hampton Research) at 4 °C and 18 °C by using the sitting-drop vapor diffusion method as previously described [19, 20]. Two protein solution drops each with 1 μl (at 7.5 and 15 mg/mL) and 1 μL of reservoir crystallizing buffer were placed and mixed together over a well containing 120 μL reservoir solution using an MRC Maxi plate (HR3–179, Hampton Research). Crystals of SLA-2*HB01-AS64-sβ2m were obtained at 14th day by using a protein concentration of 15 mg/mL in the Index solution No. 79 (0.2 M Ammonium acetate, 25% w/v PEG 3350, 0.1 M BIS-TRIS pH 6.5) at 4 °C. Crystallizing condition is shown in Table 2.
Data collection and processing
The SLA-2*HB01-AS64-sβ2m crystal was firstly soaked in reservoir solution supplemented with 17% (v/v) glycerol as a cryoprotectant for several seconds and then flash-cooled in a nitrogen stream at − 173 °C [21]. Data collection was carried out using an ADSC Q315 CCD detector at a wavelength of 1.00000 Å. By using beam line BL17U of the Shanghai Synchrotron Radiation Facility (Shanghai, China), the crystal was collected to 2.4 Å resolution. The raw data was indexed, integrated, corrected for absorption, scaled and merged using HKL-2000 [22].
Results
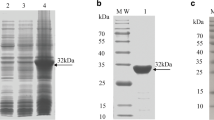
After purification to homogeneity by Superdex 200 16/60 HiLoad size-exclusion chromatography, it was shown that the heavy chain of SLA-2*HB01 was successfully refolded together with the light chain of sβ2m and the CTL epitope of AS64. The yield of the SLA-2*HB01-AS64-sβ2m complex is about 10%. It was shown four peaks in the chromatographic elution profile (Fig. 1a). The peak 1 should be aggregated heavy chains of SLA-2*HB01. The peaks 2–6 detected in lanes 2 to 6 with SDS-PAGE should be the refolded SLA-2*HB01-AS64-sβ2m complex (44 kDa). The peak 7 detected in lane 7 with SDS-PAGE should be surplus sβ2m (see inset in Fig. 1a). Another peak between peak 2–6 and peak 7, not shown in SDS-PAGE detection, should be some degraded proteins according to our previous practice. The refolded SLA-2*HB01-AS64-sβ2m complex was further purified by an anion-exchange chromatography Resource Q with a NaCl elution concentration of 21.5–26.5%. It was shown two peaks for the further purified complex (Fig. 1b). The peak 1 detected in lane 1 with SDS-PAGE contains free sβ2m. The peaks 2–4 detected in lanes 2 to 4 with SDS-PAGE contain two bands at the expected molecular weights of SLA-2*HB01 heavy chain (32 kDa) and sβ2m light chain (12 kDa) (see inset in Fig. 1b).
Purification of the refolded SLA-2*HB01-AS64-sβ2m by FPLC Superdex 200 16/60 Hi-Load gel-filtration and anion-exchange chromatography Resource Q (GE Healthcare). a Gel-filtration profile of the refolded SLA-2*HB01-AS64-sβ2m products. Peak 1 represents aggregated heavy chains of SLA-2*HB01, peaks 2–6 represent correctly refolded SLA-2*HB01-AS64-sβ2m complex (~ 44 kDa) and peak 7 represents surplus sβ2m. Inset: SDS–PAGE gel detection (15%) for peaks 1, 2–6 and 7. M, the low molecular-weight protein ladder (kDa) with 97.1, 66.3, 43.0, 31.0, 20.1 and 14.4. b Further purification of the refolded SLA-2*HB01-AS64-sβ2m by anion-exchange chromatography Resource Q. Peak 1 contains aggregated light chain of sβ2m, peaks 2–4 represent the refolded SLA-2*HB01-AS64-sβ2m products, which was eluted with a NaCl concentration of 21.5–26.5%. Inset: SDS-PAGE gel detection (15%) for peaks 1 and 2–4. M, the low molecular-weight protein ladder (kDa) with 97.1, 66.3, 43.0, 31.0, 20.1 and 14.4
The purified SLA-2*HB01-AS64-sβ2m was concentrated to 15 mg/mL for crystal screening. Crystals occurred after 7 days in the Index solution No. 79 at 4 °C and then they were harvested at 14th d (Fig. 2). After preliminary X-ray diffraction analysis, it was shown that the diffraction resolution of the SLA-2*HB01-AS64-sβ2m crystal used for data collection was 2.4 Å. The space group of the crystal belongs to P212121. The unit-cell parameters of the crystal were shown as a = 48.37, b = 97.75, c = 166.163 Å (Fig. 3). The crystal has a solvent content of 61.9%. Data-collection and processing statistics are summarized in Table 3.
Discussion
Previously, we reported a multiple amino-acid sequence alignment between SLA-2*HB01 and other known MHC class I alleles in swine, human and mouse [10]. Based on human HLA-A2 crystal data, the structure of SLA-2*HB01 was predicted by using homology modeling. We noticed SLA-2*HB01 preserved some key functional sites of HLA-A2 and H-2, which indicated that SLA-2*HB01 should be crucial in binding and presenting antigenic peptides [23, 24]. In addition, SLA-2*HB01 has 85.0–93.9, 86.2–97.0 and 83.3–88.6% sequence identity to other SLA-1, SLA-2 and SLA-3 alleles, respectively. Comprehensive analysis of SLA-2*HB01, it should be a novel allele of SLA-2 with specific genetic characteristics [10].
In this work, the crystal of the SLA-2*HB01 molecule complexed with swine β2m and a CTL epitope AS64 derived from the Asia1 serotype of FMDV was reported. It seems the space group type of SLA-2*HB01-AS64-sβ2m crystal is consistent with that of SLA-1 crystal but quite different from that of SLA-3 crystal, which indicates the structure of SLA-2 might be more similar to that of SLA-1 [17, 18]. Recently, a crystal of SLA-2*HB01 complex associated with an Hu64 CTL epitope derived from O serotype of FMDV was reported. It was shown that the two crystals were similar in space group type, but different in unit-cell parameters [19]. To learn about the elaborate structural characteristics of SLA-2, especially the special characteristics that differ from SLA-1 and SLA-3, the 3-dimentional structure of the SLA-2 complex associated more CTL epitopes derived from swine-origin virus is required to be revealed as soon as possible.
Conclusions
The research data will be used to further elucidate the 3-dimentional structure of the SLA-2 molecules and design more refined viral epitopes.
Abbreviations
- APCs:
-
Antigen presenting cells
- CTL:
-
Cytotoxic T lymphocyte antigen
- DMSO:
-
Dimethyl sulfoxide
- FMDV:
-
Foot-and-mouth disease virus
- IPTG:
-
Isopropyl-β-D-thiogalactopyranoside
- LB:
-
Luria–Bertani medium
- MHC:
-
Major histocompatibility complex
- SLA:
-
Swine leukocyte antigen
References
Lu X, Gibbs JS, Hickman HD, David A, Dolan BP, Jin Y, Kranz DM, Bennink JR, Yewdell JW, Varma R. Endogenous viral antigen processing generates peptide-specific MHC class I cell-surface clusters. Proc Natl Acad Sci U S A. 2012;109(38):15407–12.
Weidanz JA, Piazza P, Hickman-Miller H, Woodburn D, Nguyen T, Wahl A, Neethling F, Chiriva-Internati M, Rinaldo CR, Hildebrand WH. Development and implementation of a direct detection, quantitation and validation system for class I MHC self-peptide epitopes. J Immunol Methods. 2007;318(1–2):47–58.
Vigneron N, Ferrari V, Stroobant V, Abi Habib J, Van den Eynde BJ. Peptide splicing by the proteasome. J Biol Chem. 2017;292(51):21170–9.
Wolf BJ, Princiotta MF. Viral and bacterial minigene products are presented by MHC class I molecules with similar efficiencies. Mol Immunol. 2011;48(4):463–71.
Klein J, Satta Y, O'HUigin C, Takahata N. The molecular descent of the major histocompatibility complex. Annu Rev Immunol. 1993;11:269–95.
Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384(6605):134–41.
Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. J Immunol. 2005;174(1):6–19.
Smith TP, Rohrer GA, Alexander LJ, Troyer DL, Kirby-Dobbels KR, Janzen MA, Cornwell DL, Louis CF, Schook LB, Beattie CW. Directed integration of the physical and genetic linkage maps of swine chromosome 7 reveals that the SLA spans the centromere. Genome Res. 1995;5(3):259–71.
Ando A, Kawata H, Shigenari A, Anzai T, Ota M, Katsuyama Y, Sada M, Goto R, Takeshima SN, Aida Y, et al. Genetic polymorphism of the swine major histocompatibility complex ( SLA) class I genes, SLA-1, −2 and −3. Immunogenetics. 2003;55(9):583–93.
Gao FS, Bai J, Gong XJ, Zhang XH, Zhang WJ, Guo D, Zhang S. Analyzing the genetic characteristics and function of the swine leukocyte antigen 2 gene in a Chinese inbreed of pigs. Microbiol Immunol. 2012;56(3):208–15.
Salguero FJ, Sanchez-Martin MA, Diaz-San Segundo F, de Avila A, Sevilla N. Foot-and-mouth disease virus (FMDV) causes an acute disease that can be lethal for adult laboratory mice. Virology. 2005;332(1):384–96.
Tully DC, Fares MA. Unravelling selection shifts among foot-and-mouth disease virus (FMDV) serotypes. Evol Bioinformatics Online. 2006;2:211–25.
Yang J, Leen EN, Maree FF, Curry S. Crystal structure of the 3C protease from southern African territories type 2 foot-and-mouth disease virus. PeerJ. 2016;4:e1964.
Baba Sheikh MO, Rashid PMA, Marouf AS, Raheem ZH, Janga SC. Phylogenic analysis of serotype Asia1 foot-and-mouth disease virus from Sulaimani/Iraq using VP1 protein: heterogeneity with vaccine strain As1/Shamir/89. Iran J Vet Res. 2017;18(3):212–5.
Subramaniam S, Mohapatra JK, Sharma GK, Das B, Dash BB, Sanyal A, Pattnaik B. Phylogeny and genetic diversity of foot and mouth disease virus serotype Asia1 in India during 1964-2012. Vet Microbiol. 2013;167(3–4):280–8.
Pan X, Qi J, Zhang N, Li Q, Yin C, Chen R, Gao F, Xia C. Complex assembly, crystallization and preliminary X-ray crystallographic studies of the swine major histocompatibility complex molecule SLA-1*1502. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67(Pt 5):568–71.
Zhang N, Qi J, Feng S, Gao F, Liu J, Pan X, Chen R, Li Q, Chen Z, Li X, et al. Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza a H1N1 virus cytotoxic T lymphocyte epitope peptides. J Virol. 2011;85(22):11709–24.
Fan S, Wu Y, Wang S, Wang Z, Jiang B, Liu Y, Liang R, Zhou W, Zhang N, Xia C. Structural and biochemical analyses of swine major histocompatibility complex class I complexes and prediction of the epitope map of important influenza a virus strains. J Virol. 2016;90(15):6625–41.
Feng L, Sun MW, Jiang P, Li ZB, Gao H, Zhai XX, Han Y, Zhang ZH, Gao FS. Purification, crystallization and preliminary X-ray crystallographic studies of swine MHC class I complexed with an FMDV CTL epitope Hu64. Res Vet Sci. 2018;119:205–8.
Chen Z, Zhang N, Qi J, Chen R, Dijkstra JM, Li X, Wang Z, Wang J, Wu Y, Xia C. The structure of the MHC class I molecule of bony fishes provides insights into the conserved nature of the antigen-presenting system. J Immunol. 2017;199(10):3668–78.
Parkin SH, H. J Appl Crystallogr. 1998;31:945–53.
Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–26.
Gao FS, Bai J, Zhang Q, Xu CB, Li Y. Construction of multiple recombinant SLA-I proteins by linking heavy chains and light chains in vitro and analyzing their secondary and 3-dimensional structures. Gene. 2012;502(2):147–53.
Gao FS, Zhai XX, Jiang P, Zhang Q, Gao H, Li ZB, Han Y, Yang J, Zhang ZH. Identification of two novel foot-and-mouth disease virus cytotoxic T lymphocyte epitopes that can bind six SLA-I proteins. Gene. 2018;653:91–101.
Acknowledgments
We thank Professor George F. Gao (Institute of Microbiology, Chinese Academy of Sciences) for providing experimental condition.
Funding
This work was supported in part by a grant from the National Natural Science Foundation of China (31672525), a grant from the National Natural Science Foundation of China (31172304), and a grant from the Liaoning Education Bureau Key Laboratory Project (LZ2015003).
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
FSG designed, wrote, revised and submitted the manuscript. LF and PJ contributed to conduct the work, analysis and interpretation of results. ZBL, HG, XXZ and ZHZ contributed to the data collection and analysis. XH contributed to the manuscript revision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Gao, Fs., Feng, L., Jiang, P. et al. Expression, purification, crystallization and preliminary X-ray diffraction analysis of swine leukocyte antigen 2 complexed with a CTL epitope AS64 derived from Asia1 serotype of foot-and-mouth disease virus. BMC Vet Res 14, 407 (2018). https://doi.org/10.1186/s12917-018-1742-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-018-1742-3