Abstract
Background
The National Early Warning Score (NEWS2) is currently recommended in the UK for the risk stratification of COVID-19 patients, but little is known about its ability to detect severe cases. We aimed to evaluate NEWS2 for the prediction of severe COVID-19 outcome and identify and validate a set of blood and physiological parameters routinely collected at hospital admission to improve upon the use of NEWS2 alone for medium-term risk stratification.
Methods
Training cohorts comprised 1276 patients admitted to King’s College Hospital National Health Service (NHS) Foundation Trust with COVID-19 disease from 1 March to 30 April 2020. External validation cohorts included 6237 patients from five UK NHS Trusts (Guy’s and St Thomas’ Hospitals, University Hospitals Southampton, University Hospitals Bristol and Weston NHS Foundation Trust, University College London Hospitals, University Hospitals Birmingham), one hospital in Norway (Oslo University Hospital), and two hospitals in Wuhan, China (Wuhan Sixth Hospital and Taikang Tongji Hospital). The outcome was severe COVID-19 disease (transfer to intensive care unit (ICU) or death) at 14 days after hospital admission. Age, physiological measures, blood biomarkers, sex, ethnicity, and comorbidities (hypertension, diabetes, cardiovascular, respiratory and kidney diseases) measured at hospital admission were considered in the models.
Results
A baseline model of ‘NEWS2 + age’ had poor-to-moderate discrimination for severe COVID-19 infection at 14 days (area under receiver operating characteristic curve (AUC) in training cohort = 0.700, 95% confidence interval (CI) 0.680, 0.722; Brier score = 0.192, 95% CI 0.186, 0.197). A supplemented model adding eight routinely collected blood and physiological parameters (supplemental oxygen flow rate, urea, age, oxygen saturation, C-reactive protein, estimated glomerular filtration rate, neutrophil count, neutrophil/lymphocyte ratio) improved discrimination (AUC = 0.735; 95% CI 0.715, 0.757), and these improvements were replicated across seven UK and non-UK sites. However, there was evidence of miscalibration with the model tending to underestimate risks in most sites.
Conclusions
NEWS2 score had poor-to-moderate discrimination for medium-term COVID-19 outcome which raises questions about its use as a screening tool at hospital admission. Risk stratification was improved by including readily available blood and physiological parameters measured at hospital admission, but there was evidence of miscalibration in external sites. This highlights the need for a better understanding of the use of early warning scores for COVID.
Similar content being viewed by others
Key messages
-
The National Early Warning Score (NEWS2), currently recommended for stratification of severe COVID-19 disease in the UK, showed poor-to-moderate discrimination for medium-term outcomes (14-day transfer to intensive care unit (ICU) or death) amongst COVID-19 patients.
-
Risk stratification was improved by the addition of routinely measured blood and physiological parameters routinely at hospital admission (supplemental oxygen, urea, oxygen saturation, C-reactive protein, estimated glomerular filtration rate, neutrophil count, neutrophil/lymphocyte ratio) which provided moderate improvements in a risk stratification model for 14-day ICU/death.
-
This improvement over NEWS2 alone was maintained across multiple hospital trusts, but the model tended to be miscalibrated with risks of severe outcomes underestimated in most sites.
-
We benefited from existing pipelines for informatics at King’s College Hospital such as CogStack that allowed rapid extraction and processing of electronic health records. This methodological approach provided rapid insights and allowed us to overcome the complications associated with slow data centralisation approaches.
Background
As of 9 December 2020, there have been > 67 million confirmed cases of COVID-19 disease worldwide [1]. While approximately 80% of infected individuals have mild or no symptoms [2], some develop severe COVID-19 disease requiring hospital admission. Within the subset of those requiring hospitalisation, early identification of those who deteriorate and require transfer to an intensive care unit (ICU) for organ support or may die is vital.
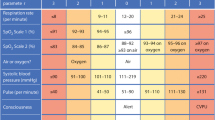
Currently, available risk scores for deterioration of acutely ill patients include (i) widely used generic ward-based risk indices such as the National Early Warning Score (NEWS2, [3]), (ii) the Modified Sequential Organ Failure Assessment (mSOFA) [4] and Quick Sequential Organ Failure Assessment [5] scoring systems, and (iii) the pneumonia-specific risk index, CURB-65 [6] which combines physiological observations with limited blood markers and comorbidities. NEWS2 is a summary score of six physiological parameters or ‘vital signs’ (respiratory rate, oxygen saturation, systolic blood pressure, heart rate, level of consciousness, temperature and supplemental oxygen dependency) used to identify patients at risk of early clinical deterioration in the United Kingdom (UK) National Health Service (NHS) hospitals [7, 8] and primary care. Some components (in particular, patient temperature, oxygen saturation, and supplemental oxygen dependency) have been associated with COVID-19 outcomes [2], but little is known about their predictive value for COVID-19 disease severity in hospitalised patients [9]. Additionally, a number of COVID-19-specific risk indices are being developed [10, 11] as well as unvalidated online calculators [12], but generalisability is unknown [13]. A Chinese study has suggested a modified version of NEWS2 with the addition of age only [14] but without any data on performance. With near-universal usage of NEWS2 in UK NHS Trusts since March 2019 [15], a minor adaptation to NEWS2 would be relatively easy to implement.
As the SARS-Cov2 pandemic has progressed, a number of risk prediction models to support clinical decisions, triage, and care in hospitalised patients have been proposed [13] incorporating potentially useful blood biomarkers [2, 16,17,18,19]. These include neutrophilia and lymphopenia, particularly in older adults [11, 18, 20, 21]; neutrophil-to-lymphocyte ratio [22]; C-reactive protein (CRP) [13]; lymphocyte-to-CRP ratio [22]; markers of liver and cardiac injury such as alanine aminotransferase (ALT), aspartate aminotransferase (AST), and cardiac troponin [23]; and elevated d-dimers, ferritin and fibrinogen [2, 6, 8].
Our aim is to evaluate the NEWS2 score and identify which clinical and blood biomarkers routinely measured at hospital admission can improve medium-term risk stratification of severe COVID-19 outcome at 14 days from hospital admission. Our specific objectives were as follows:
-
1.
To explore independent associations of routinely measured physiological and blood parameters (including NEWS2 parameters) at hospital admission with disease severity (ICU admission or death at 14 days from hospital admission), adjusting for demographics and comorbidities
-
2.
To develop a prediction model for severe COVID-19 outcomes at 14 days combining multiple blood and physiological parameters
-
3.
To compare the discrimination, calibration, and clinical utility of the resulting model with NEWS2 score and age alone using (i) internal validation and (ii) external validation at seven UK and international sites
A recent systematic review found that most existing prediction models for COVID-19 had a high risk of bias due to non-representative samples, model overfitting, or poor reporting [13]. The analyses presented here build upon our earlier work [24] which suggested that adding age and common blood biomarkers to the NEWS2 score could improve risk stratification in patients hospitalised with COVID-19. While incorporating external validation, this preliminary work was limited in that the training sample comprised 439 patients (the cohort available at the time of model development). In the present study, we (i) expand the cohort used for model development to all 1276 patients at King’s College Hospital (KCH), (ii) use hospital admission (rather than symptom onset) as the index date, (iii) consider shorter-term outcomes (3-day ICU/death), (iv) improve the reporting of model calibration and clinical utility, and (v) increase the number of external sites from three to seven.
Methods
Study cohorts
The KCH training cohort (n = 1276) was defined as all adult inpatients testing positive for severe acute respiratory syndrome coronavirus 2 (SARS-Cov2) by reverse transcription polymerase chain reaction (RT-PCR) between 1 March and 31 April 2020 at two acute hospitals (King’s College Hospital and Princess Royal University Hospital) in South East London (UK) of Kings College Hospital NHS Foundation Trust (KCH). All patients included in the study had symptoms consistent with COVID-19 (e.g. cough, fever, dyspnoea, myalgia, delirium, diarrhoea). For external validation purposes, we used seven cohorts:
-
1.
Guy’s and St Thomas’ Hospital NHS Foundation Trust (GSTT) of 988 cases (3 March 2020 to 26 August 2020)
-
2.
University Hospitals Southampton NHS Foundation Trust (UHS) of 633 cases (7 March to 6 June 2020)
-
3.
University Hospitals Bristol and Weston NHS Foundation Trust (UHBW) of 190 cases (12 March to 11 June 2020)
-
4.
University College Hospital London (UCH) of 411 cases (1 February to 30 April 2020)
-
5.
University Hospitals Birmingham (UHB) of 1037 cases (1 March to 31 June 2020)
-
6.
Oslo University Hospital (OUH) of 163 cases (6 March to 13 June 2020)
-
7.
Wuhan Sixth Hospital and Taikang Tongji Hospital of 2815 cases (4 February 2020 to 30 March 2020)
Data were extracted from structured and/or unstructured components of electronic health records (EHR) in each site as detailed below.
Measures
Outcome
For all sites, the outcome was severe COVID-19 disease at 14 days following hospital admission, categorised as transfer to the ICU/death (WHO-COVID-19 Outcomes Scales 6–8) vs. not transferred to the ICU/death (scales 3–5) [25]. For nosocomial patients (patients with symptom onset after hospital admission), the endpoint was defined as 14 days after symptom onset. Dates of hospital admission, symptom onset, ICU transfer, and death were extracted from electronic health records or ascertained manually by a clinician.
Blood and physiological parameters
We included blood and physiological parameters that were routinely obtained at hospital admission and which are routinely available in a wide range of national and international hospital and community settings. Measures available for fewer than 30% of patients were not considered (including Troponin-T, Ferritin, d-dimers and glycated haemoglobin (HbA1c), Glasgow Coma Scale score). We excluded creatinine since this parameter correlates highly (r > 0.8) with, and is used in the derivation of, estimated glomerular filtration rate. We excluded white blood cell count (WBCs) which is highly correlated with neutrophil and lymphocyte counts.
The candidate blood parameters therefore comprised albumin (g/L), C-reactive protein (CRP; mg/L), estimated glomerular filtration rate (GFR; mL/min), haemoglobin (g/L), lymphocyte count (× 109/L), neutrophil count (× 109/L), platelet count (PLT; × 109/L), neutrophil-to-lymphocyte ratio (NLR), lymphocyte-to-CRP ratio [22], and urea (mmol/L). The candidate physiological parameters included the NEWS2 total score, as well as the following parameters: respiratory rate (breaths per minute), oxygen saturation (%), supplemental oxygen flow rate (L/min), diastolic blood pressure (mmHg), systolic blood pressure (mmHg), heart rate (beats/min), and temperature (°C). For all parameters, we used the first available measure up to 48 h following hospital admission.
Demographics and comorbidities
Age, sex, ethnicity and comorbidities were considered. Self-defined ethnicity was categorised as White vs. non-White (Black, Asian, or other minority ethnic) and patients with ethnicity recorded as ‘unknown/mixed/other’ were excluded (n = 316; 25%). Binary variables were derived for comorbidities: hypertension, diabetes, heart disease (heart failure and ischemic heart disease), respiratory disease (asthma and chronic obstructive pulmonary disease (COPD)), and chronic kidney disease.
Data processing
King’s College Hospital
Data were extracted from the structured and unstructured components of the electronic health record (EHR) using natural language processing (NLP) tools belonging to the CogStack ecosystem [26], namely MedCAT [27] and MedCATTrainer [28]. The CogStack NLP pipeline captures negation, synonyms, and acronyms for medical Systematised Nomenclature of Medicine Clinical Terms (SNOMED-CT) concepts as well as surrounding linguistic context using deep learning and long short-term memory networks. MedCAT produces unsupervised annotations for all SNOMED-CT concepts (Additional file 1: Table S1) under parent terms Clinical Finding, Disorder, Organism, and Event with disambiguation, pre-trained on MIMIC-III [29]. Starting from our previous model [30], further supervised training improved detection of annotations and meta-annotations such as experiencer (is the annotated concept experienced by the patient or other), negation (is the concept annotated negated or not), and temporality (is the concept annotated in the past or present) with MedCATTrainer. Meta-annotations for hypothetical, historical, and experiencer were merged into “Irrelevant” allowing us to exclude any mentions of a concept that did not directly relate to the patient currently. Performance of the NLP pipeline for comorbidities mentioned in the text was evaluated on 4343 annotations in 146 clinical documents by a clinician (JT). F1 scores, precision, and recall are presented in Additional file 2: Table S2.
Guy’s and St Thomas’ NHS Foundation Trust
Electronic health records from all patients admitted to Guy’s and St Thomas’ NHS Foundation Trust who had a positive COVID-19 test result between 3 March and 21 May 2020, inclusive, were identified. Data were extracted using structured queries from six complementary platforms and linked using unique patient identifiers. Data processing was performed using Python 3.7 [31]. The process and outputs were reviewed by a study clinician.
University Hospitals Southampton
Data were extracted from the structured components of the UHS CHARTS EHR system and data warehouse. Data were transformed into the required format for validation purposes using Python 3.7 [31]. Diagnosis and comorbidity data of interest were gathered from the International Statistical Classification of Diseases (ICD-10) coded data. No unstructured data extraction was required for validation purposes. The process and outputs were reviewed by an experienced clinician prior to analysis.
University Hospitals Bristol and Weston NHS Foundation Trust
Data were extracted from UHBW electronic health records system (Medway). ICD-10 codes were used for diagnosis and comorbidity data. Data were transformed in line with project specifications and exported for analysis in Python 3.7 [31].
University College Hospital London
Dates of hospital admission, symptom onset, ICU transfer, and death were extracted from electronic health records. The outcome (14-day ICU/death) was defined in UCLH as ‘initiation of ventilatory support (continuous positive airway pressure, non-invasive ventilation, high-flow nasal cannula oxygen, invasive mechanical ventilation, or extracorporeal membrane oxygenation) or death’ which is consistent WHO-COVID-19 Outcomes Scales 6–8.
Wuhan cohort
Demographic, premorbid conditions, clinical symptoms or signs at presentation, laboratory data, and treatment and outcome data were extracted from electronic medical records using a standardised data collection form by a team of experienced respiratory clinicians, with double data checking and involvement of a third reviewer where there was disagreement. Anonymised data was entered into a password-protected computerised database.
University Hospitals Birmingham
Dates of hospital admission, symptom onset, ICU transfer, and death were extracted from electronic health records using the Prescribing Information and Communications System (PICS) system. The extracted data was transformed into the required format for validation purposes using Python 3.8 [31]. Diagnosis and comorbidity data of interest were gathered from ICD-10 coded data. The outcomes (3- and 14-day ICU/death) were defined consistent with WHO-COVID-19 Outcomes Scales 6–8.
Oslo University Hospital
All admitted patients with confirmed COVID-19 by positive SARS-CoV2 PCR were included in a quality registry. Data input into the register was manual. Register data was supplemented with test results from the laboratory information system (LIS) by matching exported Excel files from the register with exported Excel files from LIS. The fidelity of the match was checked against the original data source manually for a small number of patients. Only patients with symptoms consistent with COVID-19 were included in the study.
Statistical analyses
All continuous parameters were winsorized (at 1% and 99%) and scaled (mean = 0; standard deviation = 1) to facilitate interpretability and comparability [32]. Logarithmic or square root transformations were applied to skewed parameters. To explore independent associations of blood and physiological parameters with 14-day ICU/death (objective 1), we used logistic regression with Firth’s bias reduction method [33]. Each parameter was tested independently, adjusted for age and sex (model 1), and then additionally adjusted for comorbidities (model 2). P values were adjusted using the Benjamini-Hochberg procedure to keep the false discovery rate (FDR) at 5% [34].
To evaluate NEWS2 and identify parameters that could improve prediction of severe COVID-19 outcomes (objectives 2 and 3), we used regularised logistic regression with a least absolute shrinkage and selection operator (LASSO) estimator that shrinks parameters according to their variance, reduces overfitting, and enables automatic variable selection [35]. The optimal degree of regularisation was determined by identifying a tuning parameter λ using cross-validation. To avoid overfitting and to reduce the number of false-positive predictors, λ was selected to give a model with an area under the receiver operating characteristic curve (AUC) one standard error below the ‘best’ model. To evaluate the predictive performance of our model on new cases of the same underlying population (internal validation), we performed nested cross-validation (10-folds the for inner loop; 10-folds/1000 repeats for the outer loop). Discrimination was assessed using AUC and Brier score. Missing feature information was imputed using k-nearest neighbour (kNN) imputation (k = 5). All steps (feature selection, winsorizing, scaling, and kNN imputation) were incorporated within the model development and selection process to avoid data leakage that would otherwise result in optimistic performance measures [36]. All analyses were conducted with Python 3.8 [31] using the statsmodels [37] and Scikit-Learn [38] packages.
We evaluated the transportability of the derived regularised logistic regression model in external validation samples from GSTT (n = 988), UHS (n = 633), UHBW (n = 190), UCH (n = 411), UHB (n = 1037), OUH (n = 163), and Wuhan (n = 2815). Validation used LASSO logistic regression models trained on the KCH training sample, with code and pre-trained models shared via GitHub.Footnote 1 Models were assessed in terms of discrimination (AUC, sensitivity, specificity, Brier score), calibration, and clinical utility (decision curve analysis, number needed to evaluate) [32, 39]. Moderate calibration was assessed by plotting model-predicted probabilities (x-axis) against observed proportions (y-axis) with locally estimated scatterplot smoothing (LOESS) and logistic curves [40]. Clinical utility was assessed using decision curve analysis where ‘net benefit’ was plotted against a range of threshold probabilities. Unlike diagnostic performance measures, decision curves incorporate preferences of the clinician and patient. The threshold probability (pt) is where the expected benefit of treatment is equal to the expected benefit of avoiding treatment [41]. Net benefit was calculated by counting the number of true positives (predicted risk > pt and experienced severe COVID-19 outcome) and false positives (predicted risk > pt but did not experience severe COVID-19 outcome) and using the below formula:
Our model was developed as a screening tool, to identify at hospital admission patients at risk of more severe outcomes. The intended treatment for patients with a positive result from this model would be further examination by a clinician, who would make recommendations regarding appropriate treatment (e.g. earlier transfer to the ICU, intensive monitoring, treatment). We compared the decision curve from our model to two extreme cases of ‘treat none’ and ‘treat all’. The ‘treat none’ (i.e. routine management) strategy implies that no patients would be selected for further examination by a clinician; the ‘treat all’ strategy (i.e. intensive management) implies that all patients would undergo further assessment. A model is clinically beneficial if the model-implied net benefit is greater than either the ‘treat none’ or ‘treat all’ strategies.
Since the intended strategy involves a further examination by a clinician, and is therefore low risk, our emphasis throughout is on avoiding false negatives (i.e. failing to detect a severe case) at the expense of false positives. We therefore used thresholds of 30% and 20% (for 14-day and 3-day outcomes, respectively) to calculate sensitivity and specificity. This gave a better balance of sensitivity vs. specificity and reflected the clinical preference to avoid false negatives for the proposed screening tool.
Sensitivity analyses
We conducted five sensitivity analyses. First, to explore the ability of NEWS2 to predict shorter-term severe COVID-19 outcome, we developed models for ICU transfer/death at 3 days following hospital admission. All steps described above were repeated, including training (feature selection) and external validation. Second, following recent studies suggesting sex differences in COVID-19 outcome [18], we tested interactions between each physiological and blood parameters and sex using likelihood-ratio tests. Third, we repeated all models with adjustment for ethnicity in the subset of individuals with available data for ethnicity (n = 960 in the KCH training sample). Fourth, to explore the differences between community-acquired vs. nosocomial infection, we repeated all models after excluding 153 nosocomial patients (n = 1123). Finally, we considered an alternative baseline model of ‘NEWS2 only’. Our primary analyses used a baseline model of ‘NEWS2 + age’ because NEWS2 is rarely used in isolation for prognostication and treatment decisions will incorporate other patient characteristics such as age.
Results
Descriptive analyses
The KCH training cohort comprised 1276 patients admitted with a confirmed diagnosis of COVID-19 (from 1 March to 31 April 2020) of whom 389 (31%) were transferred to the ICU or died within 14 days of hospital admission, respectively. The validation cohorts comprised 6237 patients across seven sites. At UK NHS trusts, 30 to 42% of patients were transferred to the ICU or died within 14 days of admission. Disease severity was lower in the Wuhan sample, where 4% were transferred to the ICU or died. Table 1 presents the demographic and clinical characteristics of the training and validation cohorts. The UK sites were similar in terms of age and sex, with patients tending to be older (median age 59–74) and male (58 to 63%) but varied in the proportion of patients of non-White ethnicity (from 10% at UHS to 40% at KCH and UCH). Blood and physiological parameters were broadly consistent across UK sites.
Logistic regression models were used to assess independent associations between each variable and severe COVID-19 outcome (ICU transfer/death) in the KCH cohort. Additional file 3: Table S3 presents odds ratios adjusted for age and sex (model 1) and comorbidities (model 2), sorted by effect size. Increased odds of transfer to the ICU or death by 14 days were associated with NEWS2 score, oxygen flow rate, respiratory rate, CRP, neutrophil count, urea, neutrophil/lymphocyte ratio, heart rate, and temperature. Reduced odds of severe outcomes were associated with lymphocyte/CRP ratio, oxygen saturation, estimated GFR, and albumin.
Evaluating NEWS2 score for prediction of severe COVID-19 outcome
Logistic regression models were used to evaluate a baseline model containing hospital admission NEWS2 score and age for the prediction of severe COVID-19 outcomes at 14 days. Internally validated discrimination for the KCH training sample was moderate (AUC = 0.700; 95% confidence interval (CI) 0.680, 0.722; Brier score = 0.192; 0.186, 0.197; Table 2). Discrimination remained poor-to-moderate in UK validation sites (AUC = 0.623 to 0.729) but was moderate-to-good in Norway (AUC = 0.786) and Wuhan hospitals (AUC = 0.815) (Figs. 1 and 2). Calibration was inconsistent with risks underestimated in some sites (UHS, GSTT) and overestimated in others (UHBW, UHB; Fig. 2).
Supplementing NEWS2 with routinely collected blood and physiological parameters
We considered whether routine blood and physiological parameters could improve risk stratification for medium-term COVID-19 outcome (ICU transfer/death at 14 days). When adding demographic, blood, and physiological parameters to NEWS2, nine features were retained following LASSO regularisation, in order of effect size: NEWS2 score, supplemental oxygen flow rate, urea, age, oxygen saturation, CRP, estimated GFR, neutrophil count, and neutrophil/lymphocyte ratio. Notably, comorbid conditions were not retained when added in subsequent models, suggesting most of the variance explained was already captured by the included parameters. Internally validated discrimination in the KCH training sample was moderate (AUC = 0.735; 95% CI 0.715, 0.757) but improved compared to ‘NEWS2 + age’ (Table 2). This improvement over NEWS2 alone was replicated in validation samples (Fig. 1). The supplemented model continued to show evidence of substantial miscalibration.
Sensitivity analyses
For the 3-day endpoint, 13% of patients at KCH (n = 163) and between 16 and 29% of patients in the UK and Norway were transferred to the ICU or died (Table 1). The 3-day model retained just two parameters following regularisation: NEWS2 score and supplemental oxygen flow rate. For the baseline model (‘NEWS2 + age’), discrimination was moderate at internal validation (AUC = 0.764; 95% CI 0.737, 0.794; Additional file 4: Table S4) and external validation (AUC = 0.673 to 0.755), but calibration remained poor (Additional file 5: Figure S1). Moreover, the supplemented model (‘NEWS2 + oxygen flow rate’) showed smaller improvements in discrimination compared to those seen at 14 days. For the KCH training cohort, internally validated AUC increased by 0.025: from 0.764 (95% CI 0.737, 0.794) for ‘NEWS2 + age’ to 0.789 (0.763, 0.819) for the supplemented model (‘NEWS2 + oxygen flow rate’). At external validation, improvements were modest (UHBW, OUH) or negative (GSTT) in some sites, but more substantial in others (UHS, UCH). Moreover, model calibration was considerably worse for the supplemented 3-day model (Additional file 5: Figure S1).
We found no evidence of difference by sex (results not shown) and the findings were consistent when additionally adjusting for ethnicity in the subset of individuals with ethnicity data and when excluding nosocomial patients (Additional file 6: Table S5). Discrimination for the alternative baseline model of ‘NEWS2 only’ (Additional file 7: Table S6) showed a similar pattern of results as those for ‘NEWS2 + age’, except that improvements in discrimination for the supplemented model (‘All features’) were larger in most sites.
Decision curve analysis
Decision curve analysis for the 14-day endpoint is presented in Fig. 3. At KCH, the baseline model (‘NEWS2 + age’) offered small increments in net benefit compared to the ‘treat all’ and ‘treat none’ strategies for risk thresholds in the range 25 to 60%. This was replicated in all validation cohorts except for UHBW and OUH where the net benefit for ‘NEWS2 + age’ was lower than the ‘treat none’ strategy beyond the 40% risk threshold. The supplemented model (‘All features’) improved upon ‘NEWS2 + age’ and the two default strategies in most sites across the range 20 to 80%, except for (i) UHBW, where ‘treat none’ was superior beyond thresholds of 55%, and (ii) GSTT, where ‘treat all’ was superior up to a threshold of 30% and no improvement was seen for the supplemented model.
For the 3-day endpoint, the improvement in net benefit for the supplemented model over the two default strategies was smaller, compared to the improvements seen at 14 days (Additional file 8: Figure S2). At three sites (UHBW, GSST, and Wuhan), neither the baseline (‘NEWS2 + age’) nor the supplemented (‘All features’) models offered any improvement over the ‘treat all’ or ‘treat none’ strategies. At KCH and UHS, net benefit for ‘NEWS2 + age’ was higher than the default strategies for a range of risk thresholds but was not increased further by the supplemented (‘NEWS2 + oxygen flow rate’) model.
Discussion
Principal findings
This study is amongst the first to systematically evaluate NEWS2 for severe COVID-19 outcome and carry out external validation at multiple international sites (five UK NHS Trusts, one hospital in Norway, and two hospitals in Wuhan, China). We found that while ‘NEWS2 + age’ had moderate discrimination for short-term COVID-19 outcome (3-day ICU transfer/death), it showed poor-to-moderate discrimination for the medium-term outcome (14-day ICU transfer/death). Thus, while NEWS2 may be effective for short-term (e.g. 24 h) prognostication, our results question its suitability as a screening tool for medium-term COVID-19 outcome. Risk stratification was improved by adding routinely collected blood and physiological parameters, and discrimination in supplemented models was moderate-to-good. However, the model showed evidence of miscalibration, with a tendency to underestimate risks in external sites. The derived model for 14-day ICU transfer/death included nine parameters: NEWS2 score, supplemental oxygen flow rate, urea, age, oxygen saturation, CRP, estimated GFR, neutrophil count, and neutrophil/lymphocyte ratio. Notably, pre-existing comorbidities did not improve risk prediction and were not retained in the final model. This was unexpected but may indicate that the effect of pre-existing health conditions could be manifest through some of the included blood or physiological markers.
Overall, this study overcomes many of the factors associated with a high risk of bias in the development of prognostic models for COVID-19 [13] and provides some evidence to support the supplementation of NEWS2 for clinical decisions with these patients.
Comparison with other studies
A systematic review of 10 prediction models for mortality in COVID-19 infection [10] found broad similarities with the features retained in our models, particularly regarding CRP and neutrophil levels. However, existing prediction models suffer several methodological weaknesses including overfitting, selection bias, and reliance on cross-sectional data without accounting for censoring. Additionally, many existing studies have relied on single-centre or ethnically homogenous Chinese cohorts, whereas the present study shows validation across multiple and diverse populations. A key strength of our study is the robust and repeated external validation across national and international sites; however, evidence of miscalibration suggests we should be cautious when attempting to generalise these findings. Future research should include larger collaborations and aim to develop ‘from onset’ population predictions.
NEWS2 is a summary score derived from six physiological parameters, including oxygen supplementation. Lack of evidence for NEWS2 use in COVID-19 especially in primary care has been highlighted [9]. The oxygen saturation component of physiological measurements added value beyond NEWS2 total score and was retained following regularisation for 14-day endpoints. This suggests some residual association over and above what is captured by the NEWS2 score and reinforces Royal College of Physicians guidance that the NEWS2 score ceilings with respect to respiratory function [42].
Cardiac disease and myocardial injury have been described in severe COVID-19 cases in China [2, 23]. In our model, blood Troponin-T, a marker of myocardial injury, had additional salient signal but was only measured in a subset of our cohort at admission, so it was excluded from our final model. This could be explored further in larger datasets.
Strengths and limitations
Our study provides a risk stratification model for which we obtained generalisable and robust results across seven national and international sites with differing geographical catchment and population characteristics. It is amongst the first to evaluate NEWS2 at hospital admission for severe COVID-19 outcome and amongst a handful to externally validate a supplemented model across multiple sites.
However, some limitations must be acknowledged. First, there are likely to be other parameters not measured in this study that could substantially improve the risk stratification model (e.g. radiological features, obesity, or comorbidity load). These parameters could be explored in future work but were not considered in the present study to avoid limiting the real-world implementation of the risk stratification model. Second, our models showed better performance in UK secondary care settings amongst populations with higher rates of severe COVID-19 disease. Therefore, further research is needed to investigate the suitability of our model for primary care settings which have a high prevalence of mild disease severities and in community settings. This would allow us to capture variability at earlier stages of the disease and trends in patients not requiring hospital admission. Third, while external validation across multiple national and international sites represents a key strength, we did not have access to individual participant data and model development was limited to a single site (KCH). Although we benefited from existing infrastructure to support rapid data analysis, we urgently need infrastructure to support data sharing between sites to address some of the limitations of the present study (e.g. miscalibration) and improve the transferability of these models. Not only would this facilitate external validation, but more importantly, it would allow multi-site prediction models to be developed using pooled, individual participant data [43]. Fourth, our analyses would have excluded patients who experienced severe COVID-19 outcome at home or at another hospital, after being discharged from a participating hospital. Fifth, our model was restricted to blood and physiological parameters measured at hospital admission. This was by design and reflected the aim of developing a screening tool for risk stratification at hospital admission. However, future studies should explore the extent to which risk stratification could be improved by incorporating repeated measures of NEWS2 and relevant biomarkers.
Conclusions
The NEWS2 early warning score is in near-universal use in UK NHS Trusts since March 2019 [15], but little is known about its use for COVID-19 patients. Here, we showed that NEWS2 and age at hospital admission had poor-to-moderate discrimination for medium-term (14-day) severe COVID-19 outcome, questioning its use as a tool to guide hospital admission. Moreover, we showed that NEWS2 discrimination could be improved by adding eight blood and physiological parameters (supplemental oxygen flow rate, urea, age, oxygen saturation, CRP, estimated GFR, neutrophil count, neutrophil/lymphocyte ratio) that are routinely collected and readily available in healthcare services. Thus, this type of model could be easily implemented in clinical practice, and predicted risk score probabilities of individual patients are easy to communicate. At the same time, although we provided some evidence of improved discrimination vs. NEWS2 and age alone, given miscalibration in external sites, our proposed model should be used as a complement and not as a replacement for clinical judgement.
Availability of data and materials
Code and pre-trained models are available at https://github.com/ewancarr/NEWS2-COVID-19 and openly shared for testing in other COVID-19 datasets.
Source text from patient records used at all sites in the study will not be available due to inability to safely fully anonymise up to the Information Commissioner Office (ICO) standards and would be likely to contain strong identifiers (e.g. names, postcodes) and highly sensitive data (e.g. diagnoses).
A subset of the KCH dataset limited to anonymisable information (e.g. only SNOMED codes and aggregated demographics) is available on request to researchers with suitable training in information governance and human confidentiality protocols subject to approval by the King’s College Hospital Information Governance committee; applications for research access should be sent to kch-tr.cogstackrequests@nhs.net. This dataset cannot be released publicly due to the risk of re-identification of such granular individual-level data, as determined by the King’s College Hospital Caldicott Guardian.
The GSTT dataset cannot be released publicly due to the risk of re-identification of such granular individual-level data, as determined by the Guy’s and St Thomas’s Trust Caldicott Guardian.
The UHS dataset cannot be released publicly due to the risk of re-identification of such granular individual-level data, as determined by the University Hospital Southampton Caldicott Guardian.
The UCH data cannot be released publicly due to conditions of regulatory approvals that preclude open access data sharing to minimise the risk of patient identification through granular individual health record data. The authors will consider specific requests for data sharing as part of academic collaborations subject to ethical approval and data transfer agreements in accordance with the GDPR regulations.
The Wuhan dataset used in the study will not be available due to the inability to fully anonymise in line with ethical requirements. Applications for research access should be sent to TS and details will be made available via https://covid.datahelps.life/prediction/.
The OUH dataset cannot be released publicly due to the risk of re-identification of such granular individual-level data.
References
WHO. WHO COVID-19 dashboard. 2020. https://who.sprinklr.com/.
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–62.
Scott LJ, Redmond NM, Tavaré A, Little H, Srivastava S, Pullyblank A. Association between National Early Warning Scores in primary care and clinical outcomes: an observational study in UK primary and secondary care. Br J Gen Pract. 2020. https://doi.org/10.3399/bjgp20X709337.
Lambden S, Laterre PF, Levy MM, Francois B. The SOFA score—development, utility and challenges of accurate assessment in clinical trials. Crit Care. 2019;23:374.
Liu S, Yao N, Qiu Y, He C. Predictive performance of SOFA and qSOFA for in-hospital mortality in severe novel coronavirus disease. Am J Emerg Med. 2020. https://doi.org/10.1016/j.ajem.2020.07.019.
Lim WS, van der Eerden MM, Laing R, Boersma WG, Karalus N, Town GI, et al. Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax. 2003;58:377–82.
Royal College of Physicians. National Early Warning Score (NEWS) 2: standardising the assessment of acute-illness severity in the NHS. Updated report of a working party. London: RCP; 2017.
Smith GB, Prytherch DR, Meredith P, Schmidt PE, Featherstone PI. The ability of the National Early Warning Score (NEWS) to discriminate patients at risk of early cardiac arrest, unanticipated intensive care unit admission, and death. Resuscitation. 2013;84:465–70.
Greenhalgh T, Treadwell J, Burrow R. NEWS (or NEWS2) score when assessing possible COVID-19 patients in primary care. Cent Evid-Based Med Nuffield Dep Prim Care Health Sci Univ Oxf. 2020;20. https://www.cebm.net/covid-19/should-we-use-the-news-or-news2-score-when-assessing-patients-with-possible-covid-19-inprimary-care/.
Ji D, Zhang D, Xu J, Chen Z, Yang T, Zhao P, et al. Prediction for progression risk in patients with COVID-19 pneumonia: the CALL score. Clin Infect Dis. https://doi.org/10.1093/cid/ciaa414.
Shi Y, Yu X, Zhao H, Wang H, Zhao R, Sheng J. Host susceptibility to severe COVID-19 and establishment of a host risk score: findings of 487 cases outside Wuhan. Crit Care. 2020;24:108.
COVIDAnalytics. https://www.covidanalytics.io/calculator. Access date 21 April 2020.
Wynants L, Calster BV, Bonten MMJ, Collins GS, Debray TPA, Vos MD, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369. https://doi.org/10.1136/bmj.m1328.
Liao X, Wang B, Kang Y. Novel coronavirus infection during the 2019–2020 epidemic: preparing intensive care units—the experience in Sichuan Province, China. Intensive Care Med. 2020;46:357–60.
NHS England » National Early Warning Score (NEWS). https://www.england.nhs.uk/ourwork/clinical-policy/sepsis/nationalearlywarningscore/. Access date 23 April 2020.
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506.
Li K, Wu J, Wu F, Guo D, Chen L, Fang Z, et al. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Invest Radiol. 2020;Publish Ahead of Print. doi:https://doi.org/10.1097/RLI.0000000000000672.
Xie J, Tong Z, Guan X, Du B, Qiu H. Clinical characteristics of patients who died of coronavirus disease 2019 in China. JAMA Netw Open. 2020;3:e205619.
Zhang J-J, Dong X, Cao Y-Y, Yuan Y-D, Yang Y-B, Yan Y-Q, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75:1730-41.
Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–8.
Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020. https://doi.org/10.1056/NEJMoa2002032.
Lagunas-Rangel FA. Neutrophil-to-lymphocyte ratio and lymphocyte-to-C-reactive protein ratio in patients with severe coronavirus disease 2019 (COVID-19): a meta-analysis. J Med Virol. 2020;92:1733-4.
Guo T, Fan Y, Chen M, Wu X, Zhang L, He T, et al. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19). JAMA Cardiol. 2020. https://doi.org/10.1001/jamacardio.2020.1017.
Carr E, Bendayan R, Bean D, Stammers M, Wang W, Zhang H, et al. Evaluation and improvement of the National Early Warning Score (NEWS2) for COVID-19: a multi-hospital study. medRxiv. 2020. https://www.medrxiv.org/content/10.1101/2020.04.24.20078006v4.article-info.
Marshall JC, Murthy S, Diaz J, Adhikari NK, Angus DC, Arabi YM, et al. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis. 2020;20:e192–7.
Jackson R, Kartoglu I, Stringer C, Gorrell G, Roberts A, Song X, et al. CogStack - experiences of deploying integrated information retrieval and extraction services in a large National Health Service Foundation Trust hospital. BMC Med Inform Decis Mak. 2018;18. https://doi.org/10.1186/s12911-018-0623-9.
Kraljevic Z, Bean D, Mascio A, Roguski L, Folarin A, Roberts A, et al. MedCAT -- Medical Concept Annotation Tool. ArXiv191210166 Cs Stat. 2019. https://arxiv.org/abs/1912.10166.
Searle T, Kraljevic Z, Bendayan R, Bean D, Dobson R. MedCATTrainer: a biomedical free text annotation interface with active learning and research use case specific customisation. ArXiv190707322 Cs. 2019. https://arxiv.org/abs/1907.07322.
Johnson AEW, Pollard TJ, Shen L, Lehman LH, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3:1–9.
Bean D, Kraljevic Z, Searle T, Bendayan R, Pickles A, Folarin A, et al. Treatment with ACE-inhibitors is associated with less severe disease with SARS-Covid-19 infection in a multi-site UK acute Hospital Trust. medRxiv. 2020;:2020.04.07.20056788. https://www.medrxiv.org/content/10.1101/2020.04.07.20056788v1.full-text.
Van Rossum G, Drake FL. Python 3 reference manual. Scotts Valley: CreateSpace; 2009.
Steyerberg E. Clinical prediction models. 2nd ed. Cham: Springer; 2019.
Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80:27–38.
Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach. J R Stat Soc Ser B-Methodol. 1995;57:289–300.
Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Ser B Methodol. 1996;58:267–88.
Kuhn M, Johnson K. Applied predictive modeling. New York: Springer; 2013.
Seabold S, Perktold J. statsmodels: econometric and statistical modeling with Python. In: 9th Python in Science conference. Austin: SciPy; 2010. http://conference.scipy.org/proceedings/scipy2010/pdfs/seabold.pdf.
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–30.
Romero-Brufau S, Huddleston JM, Escobar GJ, Liebow M. Why the C-statistic is not informative to evaluate early warning scores and what metrics to use. Crit Care Lond Engl. 2015;19:285.
Van Calster B, McLernon DJ, van Smeden M, Wynants L, Steyerberg EW, Bossuyt P, et al. Calibration: the Achilles heel of predictive analytics. BMC Med. 2019;17:230.
Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Mak. 2006;26:565–74.
NEWS2 and deterioration in COVID-19. RCP London. 2020. https://www.rcplondon.ac.uk/news/news2-and-deterioration-covid-19. Access date 24 April 2020.
Ahmed I, Debray TP, Moons KG, Riley RD. Developing and validating risk prediction models in an individual participant data meta-analysis. BMC Med Res Methodol. 2014;14:3.
Acknowledgements
This paper represents independent research part-funded by the National Institute for Health Research (NIHR) Biomedical Research Centres at South London and Maudsley NHS Foundation Trust, London AI Medical Imaging Centre for Value-Based Healthcare, and Guy’s and St Thomas’ NHS Foundation Trust, both with King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. We would also like to thank all the clinicians managing the patients, the patient experts of the KERRI committee, Professor Irene Higginson, Professor Alastair Baker, Professor Jules Wendon, Dan Persson, and Damian Lewsley for their support.
The authors acknowledge the use of the research computing facility at King’s College London, Rosalind (https://rosalind.kcl.ac.uk), which is delivered in partnership with the National Institute for Health Research (NIHR) Biomedical Research Centres at South London & Maudsley and Guy’s and St Thomas’ NHS Foundation Trusts, and part-funded by capital equipment grants from the Maudsley Charity (award 980) and Guy’s and St Thomas’ Charity (TR130505). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, King’s College London, or the Department of Health and Social Care.
GVG also acknowledges the support from the NIHR Birmingham ECMC, NIHR Birmingham SRMRC, Nanocommons H2020-EU (731032), and the NIHR Birmingham Biomedical Research Centre. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute for Health Research, the Medical Research Council, or the Department of Health. The funding organisations had no role in the design of this study, data collection, analysis or interpretation, or preparation of the manuscript and did not approve or disapprove of or delay publication of the work. Furthermore, the UHB data collection was supported by the PIONEER Acute Care Hub and HDR-UK Better Care Programme. This work uses data provided by patients and collected by the NHS as part of their care and support. We would like to acknowledge the contribution of all staff, key workers, patients, and the community who have supported our hospitals and the wider NHS at this time.
Funding
DMB is funded by a UKRI Innovation Fellowship as part of the Health Data Research UK MR/S00310X/1 (https://www.hdruk.ac.uk).
RB is funded in part by grant MR/R016372/1 for the King’s College London MRC Skills Development Fellowship programme funded by the UK Medical Research Council (MRC, https://mrc.ukri.org) and by grant IS-BRC-1215-20018 for the National Institute for Health Research (NIHR, https://www.nihr.ac.uk) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London.
RJBD is supported by the following: (1) NIHR Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, London, UK; (2) Health Data Research UK, which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation and Wellcome Trust; (3) The BigData@Heart Consortium, funded by the Innovative Medicines Initiative-2 Joint Undertaking under grant agreement No. 116074. This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA; it is chaired by DE Grobbee and SD Anker, partnering with 20 academic and industry partners and ESC; (4) the National Institute for Health Research University College London Hospitals Biomedical Research Centre; (5) the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London; (6) the UK Research and Innovation London Medical Imaging & Artificial Intelligence Centre for Value Based Healthcare; (7) the National Institute for Health Research (NIHR) Applied Research Collaboration South London (NIHR ARC South London) at King’s College Hospital NHS Foundation Trust.
KOG is supported by an MRC Clinical Training Fellowship (MR/R017751/1).
WW is supported by the Health Foundation grant.
AD and VC acknowledge the support from the National Institute for Health Research (NIHR) Applied Research Collaboration (ARC) South London at King’s College Hospital NHS Foundation Trust and the Royal College of Physicians, as well as the support from the NIHR Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. VC is additionally supported by Health Data Research UK, which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation, and Wellcome Trust.
RZ is supported by a King’s Prize Fellowship.
AS is supported by a King’s Medical Research Trust studentship.
JTHT is supported by London AI Medical Imaging Centre for Value-Based Healthcare (AI4VBH) and the National Institute for Health Research (NIHR) Applied Research Collaboration South London (NIHR ARC South London) at King’s College Hospital NHS Foundation Trust.
FB and PTTH are funded by the National Institute for Health Research (NIHR) Biomedical Research Centre, Data Sciences at University Hospital Southampton NHS Foundation Trust, and the Clinical Informatics Research Unit, University of Southampton.
JB is funded by the Clinical Informatics Research Unit, University of Southampton, and part-funded by the Global Alliance for Chronic Disease (GDAC).
A Pinto is part-funded by UHS Digital, University Hospital Southampton, Tremona Road, Southampton.
AJS is supported by a Digital Health Fellowship through Health Education England (Wessex).
HW and HZ are supported by the Medical Research Council and Health Data Research UK Grant (MR/S004149/1), Industrial Strategy Challenge Grant (MC_PC_18029), and Wellcome Institutional Translation Partnership Award (PIII054). XW is supported by the National Natural Science Foundation of China (grant number 81700006).
AMS is supported by the British Heart Foundation (CH/1999001/11735), the National Institute for Health Research (NIHR) Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London (IS-BRC-1215-20006), and the Fondation Leducq. AP is partially supported by NIHR NF-SI-0617-10120. This work was supported by the National Institute for Health Research (NIHR) University College London Hospitals (UCH) Biomedical Research Centre (BRC) Clinical and Research Informatics Unit (CRIU), NIHR Health Informatics Collaborative (HIC), and by awards establishing the Institute of Health Informatics at University College London (UCL). This work was also supported by Health Data Research UK, which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation, and the Wellcome Trust.
RKG is funded by the NIHR (DRF-2018-11-ST2-004). MN is funded by the Wellcome Trust (207511/Z/17/Z).
The work was supported by MRC Health Data Research UK (HDRUK/CFC/01), an initiative funded by the UK Research and Innovation, Department of Health and Social Care (England) and the devolved administrations, and leading medical research charities. AK is funded by a MRC Rutherford Fellowship MR/S003991/1 (as part of Health Data Research UK https://www.hdruk.ac.uk).
Author information
Authors and Affiliations
Contributions
The corresponding author, Dr. Ewan Carr, is the guarantor of the manuscript. JT, AMS, RD, EC, and RB conceived the study design and developed the study objectives. JT, RD, AF, LR, DB, ZK, TS, and AS were the leads to develop the CogStack platform. DB, ZK, TS, and AS were responsible for the data extraction and preparation. EC, RB, AP, and DS contributed to the statistical analyses. All authors contributed to the interpretation of the data. AMS, JT, KO, and RZ provided clinical input. All authors contributed to interpret the data and draft the article and provided final approval of the manuscript. DMB, ZK, AS, TS, JTHT, LR, and KN performed the data processing and software development. KOG, RZ, and JTHT performed the data validation. At GSTT, WW and WM were responsible for the data extraction and preparation. WW performed the model validation. AD and VC contributed to the interpretation of the data. At UHS, MS and FB were responsible for the data extraction and preparation. MS, HP, and AS contributed to the statistical analysis. All authors contributed to the interpretation of the data. MS and AP provided clinical input. MS and HP performed the data/model validation. At UCH, RKG and MN were responsible for the data extraction, preparation, and model validation. At UHBW, MR and MW were responsible for the data extraction and preparation. CM and CB conducted the data and model validation. For the Wuhan cohort, XZ, XW, and JS extracted the data from the EHR system. HW and HZ preprocessed the raw data and conducted the prediction model validations. BG, HW, HZ, TS, and JS interpreted the data and results. At UHB/UoB, UHB IT and AK were responsible for the data extraction and preparation. AK, LS, VRC, and GVG performed the model validation. AK, GVG, and SB contributed to the interpretation of the data. At OUH, KW, EKA, and ARH were responsible for the data extraction and preparation. AKL contributed to the statistical analysis and performed the model validation. All authors contributed to the interpretation of the data. The views expressed are those of the authors and not necessarily those of the MRC, NHS, the NIHR, or the Department of Health and Social Care. The funders of the study had no role in the study design, data collection, data analysis, data interpretation, writing of the report, or the decision to submit the article for publication.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The KCH component of the project operated under London South East Research Ethics Committee (reference 18/LO/2048) approval granted to the King’s Electronic Records Research Interface (KERRI); specific work on COVID-19 research was reviewed with expert patient input on a virtual committee with Caldicott Guardian oversight. The UHS validation was performed as part of an urgent service evaluation agreed with approval from trust research leads and the Caldicott Guardian. For UCH, ethical approval was given by East Midlands - Nottingham 2 Research Ethics Committee (REF: 20/EM/0114; IRAS: 282900). The UHB component was operated under the PIONEER Health Data Research Hub in Acute Care ethical approval provided by the East Midlands Derby REC (reference: 20/EM/0158). For UHBW, the project was considered as service evaluation by the organisational review board. Informed consent was deemed unnecessary due to the retrospective observational nature of the data. Ethical approval for GSTT was granted by the London Bromley Research Ethics Committee (reference 20/HRA/1871) to the King’s Health Partners Data Analytics and Modelling COVID-19 Group to collect clinically relevant data points from patient’s electronic health records. The Wuhan validation was approved by the Research Ethics Committee of Shanghai Dongfang Hospital and Taikang Tongji Hospital. For the OUH validation, a project protocol was approved by the Regional Ethical Committee of South-East Norway (Reference number 137045) and the OUH data protection officer (Reference number 20/08822). Informed consent in the OUH cohort was waived because of the strictly observational nature of the project.
Consent for publication
Not applicable.
Competing interests
JTHT received research support and funding from InnovateUK, Bristol-Myers-Squibb, iRhythm Technologies, and holds shares < £5000 in Glaxo Smithkline and Biogen.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
James T Teo, Ajay M Shah, and Richard J B Dobson are joint last authors.
Supplementary Information
Additional file 1: Table S1.
SNOMED terms.
Additional file 2: Table S2.
F1, precision and recall for NLP comorbidity detection.
Additional file 3: Table S3.
Logistic regression models for each blood and physiological measure tested separately in the KCH training cohort, for 14- and 3-day ICU/death.
Additional file 4: Table S4.
Internally validated discrimination for KCH training sample based on nested repeated cross-validation.
Additional file 5: Figure S1.
Calibration (logistic and LOESS curves) of supplemented NEWS2 model for 3-day ICU/death model at validation sites.
Additional file 6: Table S5.
Univariate logistic regression models for sensitivity analyses showing odds ratios of ICU/death at 3- and 14-days for subsets of the training cohort.
Additional file 7: Table S6.
Discrimination for all models in training and validation cohorts, including alternative baseline model of ‘NEWS2 only’.
Additional file 8: Figure S2.
Net benefit of supplemented NEWS2 model for 3-day ICU/death compared to default strategies (‘treat all’ and ‘treat none’) at training and validation sites.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Carr, E., Bendayan, R., Bean, D. et al. Evaluation and improvement of the National Early Warning Score (NEWS2) for COVID-19: a multi-hospital study. BMC Med 19, 23 (2021). https://doi.org/10.1186/s12916-020-01893-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12916-020-01893-3