Abstract
Background
Everolimus, an inhibitor of mammalian target of rapamycin (mTOR), has been shown to increase the efficacy of endocrine therapies in hormone receptor (HR)-positive metastatic breast cancer. However, because breast cancer is a highly heterogeneous disease, the responses of different patients to everolimus may vary. Therefore, we performed this study to better select patients who will benefit most from or be resistant to everolimus.
Methods
Patients with HR-positive breast cancer who were treated with everolimus at the Cancer Hospital, Chinese Academy of Medical Sciences from February 2014 to March 2017 were enrolled in the present study. Mutations in ctDNA were assayed in 1021 tumor-related genes via gene panel target capture-based next-generation sequencing.
Results
In total, 120 patients with metastatic breast cancer who were treated with everolimus were enrolled in the present study. The median progression-free survival (PFS) of all patients was 5.1 months (95% confidence interval [CI] 3.9–6.3 months). No difference in survival was observed between patients who received endocrine drugs used in previous treatment regimens and patients who did not receive these drugs (median PFS 5.2 and 5.1 months, respectively, p > 0.05). Additionally, we did not find any difference in outcomes between patients who had primary resistance to previously used endocrine drugs and patients who had nonprimary resistance to previous treatments (p > 0.05). Multivariate analysis showed that < 3 metastatic sites, < 2 lines of previous endocrine therapy, < 2 lines of previous chemotherapy, and treatment with everolimus combined with fulvestrant were associated with improved survival (p < 0.05). Sixteen patients underwent ctDNA analysis before everolimus treatment. The frequency of PIK3CA gene mutations was 62.5%, and H1047R was the most frequently detected mutation. Patients with the PIK3CA/H1047R mutation had longer PFS than patients with wild-type or other mutant forms of PIK3CA, and the median PFS in these two groups of patients was 8.8 and 4.1 months, respectively (p < 0.05).
Conclusions
Our data suggest that patients who receive more lines of chemotherapy or endocrine therapy are less likely to benefit from everolimus. For everolimus combination therapy, we can even select endocrine drugs that gave rise to primary resistance in previous treatments. Additionally, the PIK3CA/H1047R mutation may be a potential biomarker of sensitivity to everolimus.
Similar content being viewed by others
Background
Hormone receptors (HRs) including estrogen receptor (ER) and/or progesterone receptor are expressed in approximately 70% of breast cancers. Endocrine therapy is a very important treatment for patients with HR-positive advanced breast cancer. However, resistance to endocrine therapy is an immense challenge in clinical settings [1, 2]. One mechanism of drug resistance to endocrine therapy is the activation of the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway [3]. The inhibitors of the PI3K/AKT/mTOR pathway could reverse endocrine drug resistance [3, 4]. Everolimus is a selective inhibitor of mTOR that has shown potential for overcoming the resistance to endocrine therapy [5,6,7]. However, due to the heterogeneity of breast cancer, a subset of patients do not respond to everolimus. Therefore, it is crucial to find biomarkers that predict the efficacy of everolimus in clinical settings [8]. Several experimental studies have indicated that cancers with PIK3CA/PTEN mutations are sensitive to everolimus; however, clinical trials did not draw the same conclusions [9,10,11,12,13,14,15,16].
To better select patients who will benefit most from or be resistant to everolimus, we conducted a retrospective analysis on data from 120 patients with metastatic breast cancer who underwent therapy at the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College from February 2014 to March 2017. We also performed circulating tumor DNA (ctDNA) analysis on sixteen patients to determine the association between gene mutations and response to everolimus.
Methods
Patients and sample collection
Patients with HR-positive breast cancer who were treated with everolimus at the Cancer Hospital, Chinese Academy of Medical Sciences from February 2014 to March 2017 were enrolled in the present study. The following data were collected for each patient: age, nuclear grade, pathological type, ER, progesterone receptor, human epidermal growth factor receptor (HER2) status, number of metastatic sites, visceral metastases, previous treatment, treatment details and clinical course. Peripheral blood samples were collected from the patients who consented to participate in the ctDNA analysis.
This study was reviewed and approved by the Ethics Committee of the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. This study was performed in accordance with the Good Clinical Practice guidelines and the Declaration of Helsinki. The need for informed consent from patients who did not participate in the ctDNA analysis was waived under the approval of the institutional review board due to the retrospective study design. Written informed consent was obtained from the patients who participated in the ctDNA analysis (ref: 16–038/1117).
Adverse events (AEs) were evaluated through reexamination or telephone follow-up at least once each month. We retrospectively collected information on AEs from patients’ medical records and laboratory test results. AEs were evaluated based on the National Cancer Institute Common Terminology Criteria for Adverse Events version 4.0.
Treatment
Patients received everolimus at a dose of 10 mg/day plus endocrine therapy including exemestane, letrozole, anastrozole, fulvestrant, tamoxifen and toremifene. The dose was reduced to 5 mg/day for patients who could not tolerate 10 mg/day. Each patient also used an oral care package that prevented stomatitis. The oral care package included kangfuxinye, a pure Chinese herbal medicine extracted from the American cockroach, a special toothbrush and a user manual for the mTOR inhibitor. Treatment with everolimus was interrupted when intolerable toxicity emerged or if patients withdrew from the study. To evaluate treatment responses, computed tomography (CT) or magnetic resonance imaging (MRI) was performed every two months or whenever signs or symptoms that indicated disease progression according to Response Evaluation Criteria in Solid Tumors (RECIST) v. 1.1 were present [17].
ctDNA analysis
Peripheral blood samples were collected in Streck tubes (Streck, Omaha, NE, USA) and were centrifuged within 72 h to separate the plasma from the peripheral blood cells. QIAamp Circulating Nucleic Acid Kits (Qiagen, Hilden, Germany) were used to extract the circulating DNA (cDNA) from 0.5–2.0 mL of the plasma samples. QIAamp DNA Blood Mini Kits (Qiagen, Hilden, Germany) were used to extract genomic DNA (gDNA) from the peripheral blood cells. Both DNA extractions were performed according to the manufacturer’s protocols, and gDNA was sequenced as the normal control sample. DNA concentration was measured using a Qubit fluorometer and the Qubit dsDNA HS (High Sensitivity) Assay Kit (Invitrogen, Carlsbad, CA, USA). The size distribution of the cfDNA was assessed using an Agilent 2100 BioAnalyzer and a DNA HS kit (Agilent Technologies, Santa Clara, CA, USA) [18]. A panel of 1021 genes was assayed in the present study (Additional file 1: Table S1). cDNA and gDNA preparation, library construction, hybrid capture, and sequencing were previously described [19]. Low-quality reads and terminal adaptor sequences were filtered out of the raw data. BWA (version 0.7.12-r1039) was employed to align the clean reads to the reference human genome (hg19). Picard (version 1.98) was used to mark PCR duplicates. GATK (version 3.4–46-gbc02625) was used for realignment and recalibration. Single nucleotide variants (SNVs) were called using MuTect (version 1.1.4) and NChot [20], a software developed in-house to review hot spot variants. GATK was used to identify small insertions and deletions (indels). CONTRA (v2.0.8) was used to identify somatic copy number variants (CNVs). Significant copy number variation was expressed as the ratio of the adjusted depth between the ctDNA and the control gDNA. We verified all the final candidate variants in the Integrative Genomics Viewer (IGV).
Statistical analysis
PFS was calculated from the date of sample collection to the date of disease progression or death from any cause. Cases that did not reach an end point (progression or death events) were censored at the date of last follow-up. Kaplan-Meier survival plots were generated based on previous treatment or gene mutations, and curves were compared using log-rank tests. All statistical tests used in the present study were two-sided, and p-values below 0.05 were considered significant. All statistical analyses were performed using SPSS version 19.0 (Chicago, IL, USA).
Results
Patients
In total, 120 patients with metastatic breast cancer who were treated with everolimus combined with other endocrine therapies at the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College from February 2014 to March 2017 were enrolled in the present study. All patients enrolled in this study were diagnosed with HR-positive breast cancer; of those, 16 (13.3%) patients had HER2-positive breast cancer, and 104 (86.7%) patients had HER2-negative breast cancer. Nineteen (15.8%) patients received tamoxifen or toremifene plus everolimus; 18 (15%) patients received fulvestrant plus everolimus; 24 (20.0%) patients received a nonsteroidal aromatase inhibitor (AI) including letrozole or anastrozole plus everolimus; and 59 (49.2%) patients received exemestane plus everolimus. The median age was 52.5 years (range, 24 to 84 years). Fifty-four (45.0%) patients had visceral metastases, and 66 (55.0%) had nonvisceral metastases. Ninety-eight (81.7%) patients had received > 2 lines of previous endocrine therapies. The main clinical characteristics of all patients in the present study are outlined in Table 1.
Survival analysis
The efficacy of everolimus was evaluated in all 120 patients. None of the patients achieved complete response (CR), but partial response (PR) was observed in 19 (15.8%) patients. Additionally, 59 (49.2%) patients showed stable disease (SD) as an optimal efficacy, and 42 (35.0%) patients showed progressive disease (PD) at first assessment. The objective response rate (ORR = CR + PR) and clinical benefit rate (CBR = CR + PR + SD) in the 120 patients were 15.8 and 65.0%, respectively. The median PFS of all patients in the present study was 5.1 months (95% confidence interval [CI] 3.894–6.306 months, Fig. 1). Overall, 39 (32.5%) patients received everolimus combined with endocrine therapies that had been used in previous treatments, and 18 (46.2%) of these patients had primary resistance against those drugs. We further analyzed the relationship between PFS and previous treatments and found no difference between patients who received endocrine drugs used in previous treatment and patients who did not receive these drugs (median PFS 5.2 and 5.1 months, respectively, p > 0.05, Fig. 2). Additionally, we did not find any difference in outcomes between patients who had primary resistance to previously received endocrine drugs and patients who had nonprimary resistance to previous treatments (p > 0.05, Fig. 3).
The PFS of patients treated with everolimus combined with endocrine therapy stratified by drugs that were previously used or not. A total of 120 patients received everolimus; 39 patients received a drug that was used in previous treatments (blue), and 81 patients received drugs that were not previously used (red). PFS, progression-free survival
The PFS of patients treated with everolimus combined with endocrine therapy stratified by primary resistance or nonprimary resistance to drugs used in previous treatments. A total of 120 patients received everolimus; 19 patients received drugs to which they acquired primary resistance during previous treatment (blue); 20 patients received drugs to which they acquired nonprimary resistance during previous treatment (red); and 81 patients received drugs that were not previously used (green). PFS, progression-free survival
Safety and tolerability
All 120 patients who received everolimus treatment were eligible for safety evaluation. The grades of all major treatment-related AEs are shown in Table 2. The most common AEs were stomatitis (23.3%) and rash (8.3%). Twenty-three patients experienced grade 3 AEs, and the main grade 3 or 4 AEs were stomatitis (9.2%), increased alanine aminotransferase (ALT) or aspartate aminotransferase (AST) levels (5.0%), cough (5.0%), and anorexia (4.2%). No treatment-related death was observed in the present study.
Risk factors for PFS
Based on the univariate analysis, patients treated with < 2 lines of previous endocrine therapy had better PFS than patients treated with > 2 lines of previous endocrine therapy (hazard ratio = 0.46, 95% CI 0.28–0.79, p = 0.005). Similarly, treatment with < 3 lines of previous chemotherapy was associated with better survival than treatment with > 3 lines of previous chemotherapy (hazard ratio = 0.48, 95% CI 0.32–0.72; p < 0.001). Moreover, patients with < 3 metastatic sites had better survival than patients with > 3 metastatic sites (hazard ratio = 0.41, 95% CI 0.27–0.62; p<0.001). In the multivariate analysis, we included the HER2 status, the number of previous endocrine therapies and chemotherapy lines, and drugs combined with everolimus. The multivariate analysis showed that improved survival was associated with < 3 metastatic sites (hazard ratio = 0.49, 95% CI 0.31–0.76; p = 0.002), < 2 lines of previous endocrine therapy (hazard ratio = 0.56, 95% CI 0.31–1.00, p = 0.048), < 2 lines of previous chemotherapy (hazard ratio = 0.59, 95% CI 0.38–0.92; p = 0.021), and treatment with everolimus combined with fulvestrant (hazard ratio = 0.53, 95% CI 0.28–0.99; p = 0.045).
ctDNA analysis
Sixteen patients underwent ctDNA analysis before everolimus treatment. Somatic genomic alterations, including CNVs and point mutations, were detected in the ctDNA of all 16 patients (100%) (Additional file 2: Table S2). The number of somatic mutations in each patient ranged from 2 to 15, and the mean number was 7.2. The commonly mutated genes were PIK3CA, TP53, ESR1, ERBB2 and ALK (Fig. 4).
The spectrum of hotspot mutations in HR-positive breast cancer patients. Each of the 18 hotspot gene mutations has been identified in more than 2 patients listed to the left of the figure. The number of mutations in each gene among the 16 patients is shown (rows). The presence of point mutations, CNVs and both are indicated in red, blue and green, respectively
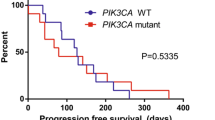
PIK3CA gene mutations were detected in 10 (62.5%) patients and included 10 point mutations and 1 CNV. PIK3CA/H1047R was the most frequently detected mutation, as it appeared in 6 patients, and the other four types of mutations, namely, H1047L, H1047Y, N345K, and E545K, were each detected in 1 patient. Patients with the PIK3CA/H1047R mutation had longer PFS than patients with wild-type or other mutant forms of PIK3CA, and the median PFS was 8.8 and 4.1 months, respectively (p = 0.020, Fig. 5). However, we did not find similar differences in survival between patients with other types of PIK3CA mutations and patients with wild-type PIK3CA in whom the median PFS was 4.6 and 7.0 months, respectively (p > 0.05).
The PFS of patients treated with everolimus based on PIK3CA/H1047R mutations. A total of 16 patients underwent ctDNA analysis; 6 patients had PIK3CA/H1047R mutations (blue), and 10 patients did not have PIK3CA/H1047R mutations (red). PFS, progression-free survival. PIK3CA, phosphatidylinositol 3-kinase
Discussion
The resistance of endocrine therapy is a major clinical challenge. The mechanism of endocrine therapy resistance is very complex, and one of the most important mechanisms is the activation of the mTOR signaling pathway [4]. Everolimus, an mTOR inhibitor, can enhance the efficacy of endocrine therapy and may reverse drug resistance [21]. We performed the present study to find patients who will benefit most from or exhibit resistance to everolimus.
In the present study, everolimus combined with endocrine therapy in patients with HR-positive metastatic breast cancer resulted in a PFS of 5.1 months. The PFS identified in the present study is shorter than the PFS in the BOLERO-2 study [5]. This disparity can be accounted for by the retrospective design of this study and the higher tumor and treatment burdens of the patients. In the present study, 45.0% patients had > 2 metastatic sites, 81.7% patients received > 2 lines of previous endocrine therapies, and 43.0% received > 2 lines of previous chemotherapy. However, in the BOLERO-2 study, only 35% of patients had > 3 metastatic sites, and 26% received chemotherapy in metastatic settings before everolimus [5, 6]. In our study, < 3 metastatic sites, < 2 lines of previous endocrine therapy, and < 2 lines of previous chemotherapy were associated with improved survival.
Additionally, the AEs profile in this study was similar to that of previous studies [5,6,7, 22]. Consistent with previous reports, stomatitis was the most frequent treatment-related adverse event. However, the frequency of stomatitis in our study was lower than that reported in previous studies. This discrepancy may be due to each patient receiving an oral care package that prevented stomatitis in our study. Effective intervention and prevention may help reduce the occurrence of severe stomatitis. Kangfuxinye has been shown to have a potential preventive effect against stomatitis and was reported to be effective in the prevention of mucositis induced by chemoradiotherapy in a phase III clinical study of nasopharyngeal carcinoma [23].
Subgroup analyses in the TAMRAD study indicated that the median time to tumor progression (TTP) was 14.8 months vs 5.4 months in patients with secondary resistance and patients with primary resistance, respectively [7]. Similarly, patients with secondary resistance to AIs had a significantly higher CBR with everolimus combined with tamoxifen (74%) than with tamoxifen alone (48%) [7]. However, in the present study, no difference in survival was observed between patients who received endocrine drugs that were used in previous treatments and patients who did not use any endocrine drugs (median PFS 5.23 and 5.10 months, respectively, p>0.05). We also did not find any difference in outcomes between patients who received endocrine drugs to which they acquired primary resistance in previous treatments and patients who received endocrine drugs to which they acquired nonprimary resistance (p > 0.05). A real-world retrospective study performed in China also found results similar to ours [22].
Thus far, no study has directly compared the efficacy among different endocrine drugs combined with everolimus. In our study, multivariate analysis indicated that treatment with everolimus combined with fulvestrant was associated with improved survival. However, these results require further confirmation in large-scale randomized clinical trials.
Mutations in the PIK3CA/AKT/mTOR pathway are frequent in breast cancer patients [24, 25]. Experimental studies have demonstrated that breast cancers with PIK3CA mutations are more sensitive to everolimus, but this has not been confirmed in clinical studies [12,13,14]. This result may be due to the heterogeneity of the mutational status between primary tumors and metastases and the small number of patients included in previous studies [8, 26]. In the present study, the frequency of PIK3CA mutations was 62.5%, which was higher than that reported in previous studies (20–45%) [19, 24]. This discrepancy may have occurred because patients in the present study had advanced cancer with increased treatment burden or because PIK3CA mutations are significantly more frequent in HR-positive cancers than in HR-negative cancers [24, 27, 28]. Based on the present study, patients with the PIK3CA/H1047R mutation had longer PFS than patients with wild-type or other mutant forms of PIK3CA, with median PFS rates of 8.8 and 4.1 months, respectively (p = 0.020). However, we did not find similar differences in survival between patients with other types of PIK3CA mutations and patients with wild-type PIK3CA in whom the median PFS was 4.6 and 7.0 months, respectively (p > 0.05). In the BOLERO-2 study, mutational analysis of plasma cell-free DNA (cfDNA) indicated no relationship between PFS after everolimus treatment and PIK3CA genotypes, which was consistent with a previous analysis of tumor tissue DNA [14]. Preclinical data suggested that PIK3CA and mTOR inhibitors had a lower IC50 for H1047R than for E542K or E545K [29]. The PIK3CA/H1047R mutation was also reported to confer sensitivity to everolimus in early-phase clinical trials in many types of cancers [30]. Therefore, PIK3CA mutations including H1047R, E545K, and E542K cannot predict patient responses to everolimus. Hence, it is possible that not all of the PIK3CA mutations confer sensitivity to everolimus and that the H1047R mutation may be a potential biomarker of sensitivity to everolimus. One possible mechanism is that the PIK3CA/H1047R mutation is a stronger driver of tumor development than other types of PIK3CA mutations such as E542K and E545K. Therefore, patients with PIK3CA/H1047R mutations were more sensitive to the mTOR inhibitor everolimus [31]. However, these results require further confirmation in clinical trials.
PTEN gene loss was another biomarker that was reported to be a marker of sensitivity to PI3K/AKT/mTOR inhibitors in preclinical studies [15, 16]. However, clinical trials including TAMRAD and BOLERO-2 did not find any association between PTEN gene status and everolimus efficacy [13, 32]. In the present study, a PTEN gene mutation was detected in only one patient, and this patient received tamoxifen plus everolimus therapy and had SD for 5.0 months. However, based on just one patient, we were unable to find an association between PTEN gene mutations in ctDNA and response to everolimus.
Despite the advantages described above, several limitations of this study should be noted. First, the retrospective design and the small sample of patients who underwent ctDNA analysis do not provide sufficient power to derive statistically sound conclusions. Additionally, because of the small sample size, we did not analyze the association between everolimus and other genetic alterations such as PTEN gene loss and KRAS gene mutations, which have been reported to be associated with everolimus response.
Conclusions
In conclusion, patients who receive more lines of chemotherapy or endocrine therapy are less likely to benefit from everolimus. Additionally, for combination with everolimus, we can also select endocrine drugs that were used or not used in previous treatments as well as drugs to which the patients had acquired primary or nonprimary resistance in previous treatments. Furthermore, the PIK3CA/H1047R mutation may be a potential biomarker of sensitivity to everolimus.
Abbreviations
- CBR:
-
Clinical benefit rate
- CNVs:
-
Copy number variants.
- CR:
-
Complete response
- CT:
-
Computed tomography
- ctDNA:
-
Circulating tumor DNA
- ER:
-
Estrogen receptor
- HER2:
-
Human epidermal growth factor receptor-2
- HR:
-
Hormone
- MRI:
-
Magnetic resonance imaging
- mTOR :
-
Mammalian target of rapamycin
- ORR:
-
Objective repose rate
- PD:
-
Progressive disease
- PFS:
-
Progression-free survival
- PI3K:
-
Phosphatidylinositol 3-kinase
- PR:
-
Partial response
- SD:
-
Stable disease
- SNVs:
-
Single nucleotide variants
References
Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45(12):1446–51.
Ma CX, Reinert T, Chmielewska I, Ellis MJ. Mechanisms of aromatase inhibitor resistance. Nat Rev Cancer. 2015;15(5):261–75.
Paplomata E, O'Regan R. The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers. Ther Adv Med Oncol. 2014;6(4):154–66.
Margariti N, Fox SB, Bottini A, Generali D. “Overcoming breast cancer drug resistance with mTOR inhibitors”. Could it be a myth or a real possibility in the short-term future? Breast Cancer Res Treat. 2011;128(3):599–606.
Yardley DA, Noguchi S, Pritchard KI, Burris HA, Baselga J, Gnant M, et al. Everolimus plus Exemestane in postmenopausal patients with HR+ breast Cancer: BOLERO-2 final progression-free survival analysis. Adv Ther. 2013;30(10):870–84.
Piccart M, Hortobagyi GN, Campone M, Pritchard KI, Lebrun F, Ito Y, et al. Everolimus plus exemestane for hormone-receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: overall survival results from BOLERO-2†. Ann Oncol. 2014;25(12):2357–62.
Bachelot T, Bourgier C, Cropet C, Ray-Coquard I, Ferrero J, Freyer G, et al. Randomized phase II trial of Everolimus in combination with tamoxifen in patients with hormone receptor–positive, human epidermal growth factor receptor 2–negative metastatic breast Cancer with prior exposure to aromatase inhibitors: A GINECO study. J Clin Oncol. 2012;30(22):2718–24.
Martelotto LG, Ng CKY, Piscuoglio S, Weigelt B, Reis-Filho JS. Breast cancer intra-tumor heterogeneity. Breast Cancer Res: BCR. 2014;16(3):210.
Wander SA, Hennessy BT, Slingerland JM. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J Clin Investig. 2011;121(4):1231–41.
Serra V, Markman B, Scaltriti M, Eichhorn PJA, Valero V, Guzman M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of Cancer cells with activating PI3K mutations. Cancer Res. 2008;68(19):8022–30.
Gonzalez-Angulo AM, Blumenschein GJ. Defining biomarkers to predict sensitivity to PI3K/Akt/mTOR pathway inhibitors in breast cancer. Cancer Treat Rev. 2013;39(4):313–20.
Janku F, Wheler JJ, Westin SN, Moulder SL, Naing A, Tsimberidou AM, et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 2012;30(8):777–82.
Hortobagyi GN, Chen D, Piccart M, Rugo HS, Burris HR, Pritchard KI, et al. Correlative analysis of genetic alterations and Everolimus benefit in hormone receptor-positive, human epidermal growth factor receptor 2-negative advanced breast Cancer: results from BOLERO-2. J Clin Oncol. 2016;34(5):419–26.
Moynahan ME, Chen D, He W, Sung P, Samoila A, You D, et al. Correlation between PIK3CA mutations in cell-free DNA and everolimus efficacy in HR+, HER2- advanced breast cancer: results from BOLERO-2. Br J Cancer. 2017;116(6):726–30.
Hong DS, Bowles DW, Falchook GS, Messersmith WA, George GC, O'Bryant CL, et al. A multicenter phase I trial of PX-866, an Oral irreversible phosphatidylinositol 3-kinase inhibitor, in patients with advanced solid tumors. Clin Cancer Res. 2012;18(15):4173–82.
O'Brien C, Wallin JJ, Sampath D, GuhaThakurta D, Savage H, Punnoose EA, et al. Predictive biomarkers of sensitivity to the phosphatidylinositol 3′ kinase inhibitor GDC-0941 in breast Cancer preclinical models. Clin Cancer Res. 2010;16(14):3670–83.
Eisenhauer EA, A PTBJ, Dancey J, G SAHS, Kaplan R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–47.
Nong J, Gong Y, Guan Y, Yi X, Yi Y, Chang L, Yang L, Lv J, Guo Z, Jia H, et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun. 2018;9(1):3114.
Yi Z, Ma F, Li C, Chen R, Yuan L, Sun X, Guan X, Li L, Liu B, Guan Y, et al. Landscape of somatic mutations in different subtypes of advanced breast cancer with circulating tumor DNA analysis. Sci Rep. 2017;7(1):5995.
Yang X, Chu Y, Zhang R, Han Y, Zhang L, Fu Y, Li D, Peng R, Li D, Ding J, et al. Technical validation of a next-generation sequencing assay for detecting clinically relevant levels of breast Cancer-related single-nucleotide variants and copy number variants using simulated cell-free DNA. J Mol Diagn. 2017;19(4):525–36.
Zagouri F, Sergentanis TN, Chrysikos D, Filipits M, Bartsch R. mTOR inhibitors in breast cancer: a systematic review. Gynecol Oncol. 2012;127(3):662–72.
Gong C, Zhao Y, Wang B, Hu X, Wang Z, Zhang J, Zhang S. Efficacy and safety of everolimus in Chinese metastatic HR positive, HER2 negative breast cancer patients: a real-world retrospective study. Oncotarget. 2017;8(35):59810–22.
Luo Y, Feng M, Fan Z, Zhu X, Jin F, Li R, Wu J, Yang X, Jiang Q, Bai H, et al. Effect of Kangfuxin solution on chemo/radiotherapy-induced mucositis in nasopharyngeal carcinoma patients: A multicenter, prospective randomized phase III clinical study. Evid Based Complement Alternat Med. 2016;2016:8692343.
Ahmad F, Badwe A, Verma G, Bhatia S, Das BR. Molecular evaluation of PIK3CA gene mutation in breast cancer: determination of frequency, distribution pattern and its association with clinicopathological findings in Indian patients. Med Oncol. 2016;33(7):74.
Villarreal-Garza C, Cortes J, Andre F, Verma S. mTOR inhibitors in the management of hormone receptor-positive breast cancer: the latest evidence and future directions. Ann Oncol. 2012;23(10):2526–35.
Amir E, Miller N, Geddie W, Freedman O, Kassam F, Simmons C, Oldfield M, Dranitsaris G, Tomlinson G, Laupacis A, et al. Prospective study evaluating the impact of tissue confirmation of metastatic disease in patients with breast cancer. J Clin Oncol. 2012;30(6):587–92.
Dirican E, Akkiprik M, Ozer A. Mutation distributions and clinical correlations of PIK3CA gene mutations in breast cancer. Tumour Biol. 2016;37(6):7033–45.
Arsenic R, Lehmann A, Budczies J, Koch I, Prinzler J, Kleine-Tebbe A, Schewe C, Loibl S, Dietel M, Denkert C. Analysis of PIK3CA mutations in breast cancer subtypes. Appl Immunohistochem Mol Morphol. 2014;22(1):50–6.
Matthews D, O Farrell M, Joyce J, Stott G, Giddens A, Rewcastle GEA. Preclinical characterization of PWT33597, a dual inhibitor of PI3-kinase alpha and mTOR. Proceedings Am Assoc Cancer Res 102nd Annual Meet. 2011:4485.
Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek VM, Fu S, Piha-Paul SA, Lee JJ, Luthra R, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73(1):276–84.
Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A. 2006;103(5):1475–9.
Treilleux I, Arnedos M, Cropet C, Wang Q, Ferrero JM, Abadie-Lacourtoisie S, Levy C, Legouffe E, Lortholary A, Pujade-Lauraine E, et al. Translational studies within the TAMRAD randomized GINECO trial: evidence for mTORC1 activation marker as a predictive factor for everolimus efficacy in advanced breast cancer. Ann Oncol. 2015;26(1):120–5.
Acknowledgments
Not applicable
Funding
The present study was supported by the National Natural Science Foundation of China (81472453), the Major Project of Beijing Municipal Science and Technology Commission (D161100000816004) and the CAMS Initiative for Innovative Medicine (CAMS-12 M-1-010, 2017-I2M-3-004). The funding bodies did not play a role in the study design, analysis, or interpretation of the data or in the writing of the manuscript or the decision to submit the manuscript for publication.
Availability of data and materials
All data generated or analyzed during this study are included in the published article, figures, tables, and additional files.
Author information
Authors and Affiliations
Contributions
FM and ZBY conceived and designed the study. XWG, LXL, BLL and BHX Provided study materials or patients. ZBY, CXL, BHX and HLQ performed the experiments. ZBY and FM analyzed the data and wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was reviewed and approved by the ethics committee at the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. The collection and sequencing of peripheral blood samples of patients who consented to participate in ctDNA analysis were approved by the ethics committee (ref: 16–038/1117). Written informed consent was obtained from all patients who participated in the ctDNA analysis. Due to the retrospective approach of this study, the need for informed consent from patients who did not participate int he ctDNA analysis was waived by the ethics committees.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. List of target region genes. (DOC 380 kb) (DOC 379 kb)
Additional file 2:
Table S2. Somatic mutations identified in 16 patients. (DOC 279 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yi, Z., Ma, F., Liu, B. et al. Everolimus in hormone receptor-positive metastatic breast cancer: PIK3CA mutation H1047R was a potential efficacy biomarker in a retrospective study. BMC Cancer 19, 442 (2019). https://doi.org/10.1186/s12885-019-5668-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-019-5668-3