Abstract
Background
Genetic alterations in pediatric primary brain tumors can be used as diagnostic and prognostic markers and are the basis for the development of new target therapies that, ideally, would be associated with lower mortality and morbidity. This study evaluates the incidence and interplay of the presence of BRAF V600E mutation and chromosomal 9p21 deletions in a series of 100 pediatric gliomas, aiming to determine the role of these alterations in recurrence and malignant transformation, and to verify if they could be used in the clinical set for stratifying patients for tailored therapies and surveillance.
Methods
Sanger sequencing was used for the assessment of BRAF mutations at exon 15 and Fluorescent In Situ Hybridization (FISH) with BAC: RP11–14192 for the detection of 9p21 alterations. Expression levels of the CDKN2A and MTAP by real-time PCR were evaluated in cases with 9p21 deletions. Statistical analysis of genetic and clinical data was performed using Graph Pad Prism 5 and SPSS Statistics 24 software.
Results
In our cohort it was observed that 7 /78 (8,9%) of the low-grade tumors recurred and 2 (2,6%) showed malignant transformation. BRAF V600E mutations were detected in 15 cases. No statistically significant correlations were found between the presence of BRAF V600E mutation and patient’s morphologic or clinical features. Deletions at 9p21 abrogating the CDKN2A/B and MTAP loci were rare in grade I gliomas (12.2%, p = 0.0178) but frequent in grade IV gliomas (62.5%, p = 0.0087). Moreover it was found that deletions at these loci were correlated with a shorter overall survival (p = 0.011) and a shorter progression-free survival (p = 0.016).
Conclusions
It was demonstrated that in these tumors BRAF V600E mutated and that CDKN2A/B MTAP co-deletions may be used for stratifying patients for a stricter surveillance. The Investigating and defining if glial tumors with CDKN2A/B and MTAP homozygous loss may be vulnerable to new forms of therapy, namely those affecting the methionine salvage pathway, was proven to be of importance.
Similar content being viewed by others
Background
Tumors of the Central Nervous System (CNS) account for 20–25% of all pediatric cancer diagnoses in the developed world, being the most prevalent group of cancers in children, after leukemia. Although uncommon, pediatric brain tumors represent the leading cause of cancer-related mortality in children and adolescents aged 20 years and under; and the third leading cause of cancer-related death in young adults aged 20 to 39 years. In addition to their usual high mortality, CNS tumors are associated with extensive morbidities such as pituitary dysfunction, growth hormone deficiency, epilepsy, vision loss, impaired motor skills, memory dysfunction, attention and behavioral disorders and reduced intelligence quotient (IQ). So, about 90% of survivors are left with long term cognitive and psycho-social deficits [1].
Glial and neuro-glial derived tumors are the most frequent CNS tumors in pediatric population [1, 2]. They form an heterogeneous group of neoplasia which are categorized by the World Health Organization (WHO) [2, 3] into various entities based on their cellular, genetic and clinical characteristics.
In what concerns their clinical features, gliomas are clustered in two major groups: low-grade gliomas (LGG) and high-grade gliomas (HGG). LGG comprise: i) the non-diffuse/non-infiltrative pilocytic astrocytoma (PA) categorized as WHO grade I tumor due to its predominantly favorable outcome, which, depending on their localization, can be cured by surgery alone: ii) the diffuse/ infiltrative gliomas which are associated with a less favorable clinical outcome, namely, recurrence after initial resection and a higher probability to malignant transformation. The most frequent histological entities ascertained to the former group are: the diffuse astrocytoma grade II, gangliogliomas, the angiocentric gliomas and pleomorphic xanthoastrocytomas.
Conversely, HGG comprise other histological entities that invariably recur and show progressive disease after the initial surgical resection. In the HGG are included all WHO grade III and IV neoplasms and the most frequent histologic categories are: anaplastic astrocytoma, anaplastic pleomorphic xanthoastrocytoma and glioblastoma (GBM).
In pediatric low-grade gliomas, the analysis of the molecular and genetic mechanisms underlying gliomagenesis allowed for an understanding of MAPK/ERK pathway activation as fundamental for their development. This occurs at high frequency by activation of the BRAF oncogene; and in lower frequencies: by MYB and MYBL1 amplifications and rearrangements, and FGFR1 rearrangements and mutations [4,5,6,7,8,9,10].
Two different mechanisms may lead to BRAF activation in pediatric brain gliomas: chromosomal rearrangements and point mutations. The most common BRAF rearrangement is the one resulting in BRAF/KIAA1549 fusion protein in which the N-terminus of the protein encoded by KIAA1549 gene is fused with the C-terminus of the protein encoded by BRAF gene, preserving the BRAF kinase domain [10, 11]. BRAF activating rearrangements were reported to be present in 70% of the pilocytic astrocytomas, in 15% of other low-grade gliomas, and have only been punctually observed in high-grade gliomas [9]. Studies performed by Hawkins et al. (2011) [12], Horbinski et al. (2010) [13], and Jones et al. (2008) [14], showed that BRAF rearrangements were an independent favorable prognostic factor in both supra-tentorial and posterior fossa low-grade gliomas.
The vast majority (> 90%) of BRAF mutations in pediatric gliomas are BRAF V600E mutations, a somatic mutation causing the substitution of the amino acid valine by glutamic acid at residue 600 of exon 15. BRAF V600E mutations have been described in a wide variety of lesions: 80% of pleomorphic xanthoastrocytomas 33% of the gangliogliomas, 23% of the diffuse astrocytomas, 10% of the glioblastomas being more frequent in tumors located in the cerebral cortex [15]. Only rarely BRAF V600E mutation occurs in conjunction with a BRAF/KIAA1549 rearrangement in the same tumor [4].
At variance with BRAF rearrangements, the role of BRAF V600E mutation in the glioma’s evolution and patient’s follow-up is far from being fully understood and some contradictory results are found in literature. Accordingly, while Horbinski et al. (2012) [13] showed that in their cohort of pediatric low-grade gliomas, BRAF V600E mutationended to a worse progression-free survival when compared to wild-type tumors, Mistry et al. (2015) [16] showed that this mutation was associated with a prolonged latency to malignant transformation and, consequently, with a better overall survival when compared to wild-type pediatric low-grade gliomas.
Moreover, Korshunov, et al. (2015) [17] described a subgroup of glioblastomas, exclusive to the pediatric population, that was characterized by the BRAF V600E mutation and CDKN2A deletion. Although these tumors had a better overall survival, they still had a high recurrence rate (67%).
The CDKN2A gene is mapped at the chromosome 9p21 region and encodes the p16INK4A and p14ARF proteins. p16INK4A protein has a key role as negative regulator of the proliferation of normal cells, controlling the progression through G1 into the S phase of the cell cycle. According to Raabe et al. (2011) [18], the worst outcomes associated with CDKN2A gene deletion could reflect a failure to induce senescence or an escape from the induced tumor senescence in BRAF driven CDKN2A− tumors.
In order to further understand the interplay between BRAF V600E mutation and the chromosomal region abrogating the CDKN2A gene: 9p21, a cohort of 100 pediatric gliomas was retrospectively analyzed.
Materials and methods
Human tumor samples
Analysis was performed in paraffin embedded material or fresh tumor samples of patients with pediatric gliomas that were referred to the Portuguese Institute of Oncology (IPOFG-Lisbon, Portugal) from 1992 to 2015 following Patient and Institutional Ethical Board Committee approval.
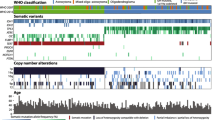
In total, 100 pediatric gliomas (PG) were selected for genetic analysis: 67 grade I gliomas, 11 grade II gliomas, 13 grade III gliomas, and 9 grade IV gliomas. Since the aim of this study was to investigate the influence of BRAF V600E mutation and 9p21 gene loss, grade I and II lesions were assembled in one group, referred as Low-Grade Gliomas (LGG) and grade III and IV in another group, referred as High-Grade Gliomas (HGG). Histological classification was performed and revised according to WHO (2007) criteria. The patients’ clinical characteristics (tumor’s location, histologic classification, WHO grade, patient’s age-ranges and gender) and outcomes are depicted in Fig. 1 and Additional file 1: Table S1. In grade I gliomas (referred in Additional file 1: Table S1 as LGG1 to LGG66, and LGG82): 32 cases were females and 35 were males, the mean age in this group was of: 9.25 years (range from 5 months to 17 years); In grade II gliomas (referred as LGG68; LGG70 to 76; LGG78, 80 and 81 in Additional file 1: Table S1): 6 cases were females, 5 males, the mean age in this group was: 6.02 years (range from 1 year to 17 years). As to grade III gliomas (referred in Additional file 1: Table S1 1 as HGG1–2; HGG5 to 15): 3 were females, 10 were males and the mean age of this group was: 9.36 years (range from 9 months to 16 years). In grade IV glioma group of this study (referred in Additional file 1: Table S1 as HGG3; HGG16 to 23): 2 patients were females and 7 males, the mean age in this group was 10.44 years (range from 5 years to 15 years).
Graphical representation of the clinical data and of the genetic alterations observed in the cohort of pediatric gliomas. Abreviations: AB (Grade IV Astroblastoma), AO (Anaplastic Oligodendroglioma), AOA (Anaplastic Oligoastrocytoma), DA (Diffuse Astrocytoma), DIG (Desmoplastic Infantile Ganglioglioma), DIPG (Diffuse Intrinsic Pontine Glioma); DNET (Dysembryoplastic Neuroepithelial Tumor), GB (Glioblastoma), GC (Gangliocytoma), GG (Ganglioglioma), GT (Glioneuronal Tumor) OG (Oligodendroglioma), PMA (Pilomyxoid Astrocytoma), PA (Pilocytic Astrocytoma), PGT (Papillary Glioneuronal Tumor), SEGA (Subependymal Giant Cell Astrocytoma), TGA (Anaplastic Glioneuronal Tumor), XA (Xanthoastrocytoma), XP (Pleomorphic Xanthoastrocytoma), AA (Anaplastic Astrocytoma), ND (Not Done). Red squares represent the presence of the genetic alteration
In accordance with WHO 2007 criteria, patients were considered to have a recurrent tumor when exhibiting a glioma of the same histologic grade after at least 1 year of remission. They were considered to have a de novo tumor when exhibiting a glioma of the same grade, with a different histological type and at a different localization. Patients were only considered as having a transformation of LGG to HGG if they exhibited one of the following features: i) consecutive histologic diagnosis of LGG and HGG as per the WHO 2007 criteria, ii) histological diagnosis LGG follow by at least 1 year of stable disease, followed by clinical and radiological progression to HGG.
Fluorescent in situ hybridization (FISH)
FISH technique was used to identify heterozygous and homozygous deletions of the 9p21 chromosomal region. It was performed in interphase nuclei with DNA probes indirectly labeled. A locus-specific BAC probe (RP11–14912) directed to the chromosomal location 9p21.3 within chromosome 9–21,909,260 and 22,010,414 (Ensembl Homo sapiens version 86.38 (GRCh38.p7) - was used to identify the 9p21 chromosomal region. An enumeration probe for chromosome 9 (pMR9A) was also used. The 9p21 chromosomal region probe was labeled with digoxigenin and the chromosome 9 probe with biotin using the BioPrimeR DNA Labeling System (Invitrogen™,by Thermo Fisher Scientific, USA). These labels were then detected with specific antibodies: anti-digoxigenin-fluorescein (Roche Diagnostics, Germany) and streptavidin combined with cyanine 3 (Cy3) fluorophore (Jackson ImmunoResearch Laboratories, USA), respectively. FISH signal evaluation was performed in an AxioImager fluorescence microscope linked to aCytoVision® software (Applied Imaging, UK). Analysis was performed by counting at least 100 cells per slide with intact, non-overlapping nuclei taken from different, randomly chosen fields of view. Deletion was defined as more than 40% containing 9p21 probe: cep9 probe ≤0.5.
Sequencing using sanger methodology
In the context of this work, sequencing using Sanger methodology was used to identify a single point mutation (V600E) in the exon 15 of the BRAF gene. DNA for gene analysis was extracted by different procedures according to the material used (fresh material or paraffin embedded material). Target gene amplification was achieved through polymerase chain reaction (PCR), using the forward primer 5’TCATAATGCTTGCTCTGATAGGA3’ and the reverse primer 5’CCGGTTTTTAAATTAGTCACCT3’ at an annealing temperature of 58–59.5 °C. To sequence samples, an automatic sequencer, ABI PrismTM 3130 Genetic Analyser (Applied Biosystems), was used. Electrophoretograms were compared with the reference DNA sequence of the studied gene to identify the presence or absence of mutations. Reference DNA sequences were obtained from Ensembl data base, available at: http://www.ensembl.org/index.html.
Expression studies using quantitative real-time RT-PCR
In cases with 9p21 deletions (detected by FISH) expression analysis was performed by real-time qPCR, using commercial TaqMan probes for the CDKN2A (Hs00233365_m1) and MTAP (Hs00559618_m1) genes (ThermoFisher Scientific, USA). Analysis was performed using QuantStudio5-Applied Biosystems Real-Time PCR System. ABL1 probes as described by Beillard E et al. [19] were used as endogenous control for gene expression assays. RNA from 3 sample cases (CN1, CN2, and CN3), defined as non-neoplastic brain lesions by pathology, were used as calibrator controls. We were not able to extract RNA with sufficient quality and purity, as analysed by agarose gel electrophoresis and Nanodrop spectrophotometry (NanoDrop Technologies,USA), from all cases with 9p21 loss of this cohort. Total RNA extraction and single-strand cDNA synthesis could only be performed for 3 LGG presenting 9p21 heterozygous loss: LGG64, LGG73 and LGG74 and for 3 HGG (grade IV) with 9p21 homozygous loss: HGG3, HGG18 and HGG23. Total RNA was extracted from 30 mg of the tumor specimens using the RNeasy® Mini Kit (Qiagen) and c-DNA synthesis was performed using the superscript II kit (Invitrogen, ThermoFisher Scientific). Gene expression was quantified on QuantStudio5 (Applied Biosystems, ThermoFisher Scientific, USA). Real-Time PCR System was performed as per manufactures protocol. CDKN2A and MTAP expression were normalized with to reference gene ABL1 and the fold change relative to CN1, CN2, CN3 samples was calculated.
Statistical analysis
Statistical analyses were carried out using GraphPad Prism 5 software (San Diego, USA). The two-tailed Fisher’s exact test was used to determine the correlations between the presence or absence of genetic alterations and patients’ gender, tumors’ WHO grade, histology and location. The Mann-Whitney test was used to study the presence or absence of genetic alterations with patient’s age. Survival curves were compared using the Log-Rank (Mantel-Cox) test using the IBM SPSS Statistics 24 Software. Differences were considered statistically significant at p-value < 0.05.
Results
BRAF V600E mutation
In total, 94 pediatric gliomas were analyzed for the BRAF V600E mutation and 15 were found to harbor this mutation (Fig. 1 and Additional file 1: Table S1): 9/62 (14,5%) grade I gliomas (6 pilocytic astrocytomas (PA) and 3 gangliogliomas (GG)); 2/11 (18,2%) grade II gliomas (1 diffuse astrocytoma (DA) and 1 pleomorphic xanthoastrocytoma (XP) classified as a grade II lesion); 2/13 (15,4%) grade III gliomas (1 anaplastic astrocytoma (AA) and 1 anaplastic XP); and 2/9 (22,2%) grade IV gliomas (one astroblastoma and one glioblastoma (GBM)).
The presence of BRAFV600E mutation did not show any correlation with tumor’s WHO grade, histologic subtype, patients’ age (p = 0.9299) and patients’ gender (p = 0.1539). This mutation was not found in tumors located in the cerebellum (p = 0.0355).
Regarding the correlation of BRAF V600E mutation with the overall survival rate of the patients, although no statistically significant correlation could be observed (p = 0,375), a trend toward a better overall survival (OS) was detected (Fig. 2). No statistically significant correlation (p = 0,299) was observed when analyzing progression free survival (PFS) differences between BRAF wild type (BRAFwt) and BRAF V600E mutated (BRAFmut) tumors (Fig. 2).
Correlation between BRAF and 9p21 alterations and the clinical outcome - Kaplan-Meier curves. a: Overall Survival (OS) curves of BRAF V600E – positive tumors v.s. BRAFwt tumors, p = 0.375; b: Progression-Free Survival (PFS) curves of BRAF V600E – positive tumors v.s. BRAFwt tumors, p = 0.299; c: OS curves of the 9p21 chromosomal region deleted tumors v.s those without 9p21 deletion, p = 0.011; (d) PFS curves of the 9p21 chromosomal region deleted tumors v.s those without 9p21 deletion, p = 0.016
9p21 chromosomal region deletion
For the 9p21 chromosomal region, 76 gliomas were analyzed, and its deletion was identified in 16 cases, as depicted in Fig. 1 and Additional file 1: Table S1. 9p21 chromosomal region deletions were observed in 10/59 (16,9%) LGG (cases LGG: 6, 12, 37, 46, 47, 64, 72 to 74 and 81) in which all deletions were heterozygous. 6/17 (35,3%) HGG (HGG: 3, 9, 18, 19, 21 and 23) presented 9p21 deletions, four of which had a homozygous 9p21 chromosomal region deletion.
Statistical analysis demonstrated that 9p21 chromosomal region deletion did not correlate with tumor location, patients’ age (p = 0.6093) and patients’ gender (p = 0.7831), but it did correlate with gliomas’ grade. The 9p21 chromosomal region deletion was rare in grade I gliomas (12.2%, p = 0.0178) but frequent in grade IV gliomas (62.5%, p = 0.0087). Moreover, this deletion also correlated with the histology categorization of tumors, a p = 0 .0012 was found between 9p21 loss and glioblastoma histology.
Furthermore, as depicted in Fig. 2 the 9p21 chromosomal region deletion was correlated with a worse overall survival rate (p = 0.011). The PFS values of 9p21 loss group were also significantly (p = 0.016) worse than those of the group with no deletion at 9p21 as observed in Fig. 2.
BRAF V600E and 9p21 deletion, in recurrence, progression and development of secondary HGG
Of the 100 gliomas evaluated in our series during a 23-year period it was observed that in the 78 LGG group, 7 recurred (8,9%) (LGG 2, 3, 34, 36, 40, 54, 68) and 2 (2,6%) showed progression to HGG (LGG81 and LGG82) (Table 1). It must be noted that the development of gliomas in the patient identified as LGG68 was very interesting. This patient (ruled out as having Neurofibromatosis type 1) was diagnosed at the age of 4 with a diffuse astrocytoma of the optical nerve, which was positive for the BRAF V600E mutation and presented a recurrence, 3 years later, that also was positive for BRAF V600E. However, at the age of 5 (two years before recurrence) he also had a pilocytic astrocytoma, which developed at the cerebral cortex that did not present BRAF V600E mutation. These findings suggest that the cortical and the optical nerve’s LGG of the patient had different cells of origin. This tumor was therefore considered as de novo glioma.
In grade III HGG group, five patients showed progressive disease and died (cases HGG1, 8, 9, 10 and 11), 4 patients were alive (HGG5, 6, 12 and 14) and in 3 cases follow-up was lost.
As for grade IV HGG group (n = 9) three patients died (HGG 3, 18, 23) of progressive disease. HGG18 was considered a secondary HGG. This patient had been diagnosed in 1998 at age of 4 with acute lymphoblastic leukemia type B (ALL-B), received a medullar transplant and was treated with radiotherapy. He was considered at remission between 2003 and 2009 but, in 2009 at the age of 15, he was diagnosed as having a glioblastoma. Of the other grade IV gliomas of our series, two (HGG15 and 16) were still alive in 2015. Both tumors were classified as astroblastomas. One, HGG15 had a BRAF V600E mutation. However, none of them presented a deletion of 9p21 region. Four cases (HGG19, 20, 21, 22) were lost for follow-up.
CDKN2A and MTAP expression studies by quantitative real-time RT-PCR
The expression levels for the CDKN2A and MTAP genes in both the LGG and HGG are depicted in Additional file 2: Figure S1 and in Additional file 3. In the LGG cases expression levels for the CDKN2A gene mRNA levels were normal or elevated. A similar situation was observed for the MTAP gene for cases LGG64 and LGG73. However, in LGG74, identified as pilomyxoid astrocytoma BRAFwt, MTAP gene levels were slightly down-regulated. In all HGG, expression levels for both genes were significantly down-regulated.
Discussion
Incidence and interplay of the presence of BRAF V600E mutation and 9p21 region deletion was evaluated in a series of 78 LGG and 22 HGG aiming to determine the role of these two alterations in recurrence and gliomas’ malignant transformation. There was also interest in verifying if they could be used in a clinical set for stratifying patients for existing target therapies and a tailored strict surveillance.
As to the incidence rate of BRAF V600E mutation results corroborated previous data [4, 15, 17], showing that this mutation occurred in all spectra of glioma types, although being more frequent in gangliogliomas and diffuse astrocytomas. Accordingly, we identified BRAF V600E mutation in 43% of the gangliogliomas, in 33% of the diffuse astrocytomas in 13% of the pilocytic astrocytomas, but also in one anaplastic astrocytoma and one glioblastoma.
In our population we observed a recurrence incidence of 8.9% and a 2,6% incidence of pediatric LGG, which undergo malignant transformation into HGG.
Statistical evaluation revealed no correlation between the presence of BRAF V600E and recurrence or progression in our series.
Mistry, et al. (2015) [16] by analysis of a cohort of 886 patients reported a similar incidence rate of pediatric LGG transformation to sHGG - 2,9%, but at variance with our data they showed BRAF V600E mutation to be associated with prolonged latency periods in pediatric low-grade gliomas. Our data suggests that tumor progression is independent of the presence of BRAF V600E mutation since progression free survival rates were very similar between the two groups: BRAFwt and BRAFmut.
In our series 9p21 deletion was rare in grade I (12.2%, p = 0.0178) but frequent in grade IV gliomas (62.5%, p = 0.0087). Moreover, this deletion also correlated with the histologic categorization of tumors, a p = 0.0012 was found between 9p21 deletion and glioblastoma histology.
In accordance with previous studies performed by Horbinski et al. (2012) [13], Mistry et al. (2015) [16] and Roy et al. (2016) [20] results led to the conclusion that homozygous or heterozygous deletions of the 9p21 chromosomal region were associated with a shorter overall survival and with a worse progression-free survival.
Interestingly, the two pediatric LGG (LGG81 and 82) that underwent malignant transformation into HGG (HGG 23 and 9 respectively), both presented a 9p21 region deletion. One, LGG81, also had a BRAF V600E mutation ab initio and the observed latency period until the development of the HGG was two years. LGG82 did not present ab initio a BRAF V600E mutation and had a much longer latency period until the development of the HGG (13 years). Although the number of cases is recognizably small, these findings suggest that in LGG81, a cooperative interplay might have occurred between BRAF mutation and 9p21 homozygous loss, which enhanced malignant transformation.
Several tumor suppressor genes (TSG) are known to be located at 9p21, namely, the Cyclin Dependent Kinase Inhibitor’s: CDKN2A and B; the Kelch Like Family Member 9 (KLHL9) and the Metylthioadenosine Phosphorilase (MTAP). The role of all these TSG in gliomas has been under investigation. Huilard et al. (2012) [21] by performing functional studies in neural progenitor cell and mouse models evidenced that homozygous CDKN2A/INK4a-Arf deletion in BRAF V600E expressing cells was sufficient for the formation of tumors with histologic features similar to the malignant astrocytomas in humans [21]. Mistry et al. (2015) [16] in an array of comparative genomic hybridization analysis of 886 pediatric gliomas alsodefined that CDKN2A deletions had a central role in low-grade glioma malignant transformation. On the other hand, Roy et al. (2016) [20] in a cohort of 379 adult LGG, conversely observed in their genomic analysis that CDKN2A inactivation was not associated with progression. Indeed, by correlating gene expression with zygosity status, they found that CDKN2A expression levels were normal or even elevated when associated at heterozygous gene loss, and proposed rather that KLHL9 and MTAP would concurrently and in a context-dependent manner promote tumor aggressiveness.
In this study BAC-RP11–14912 was used for FISH analysis. This BAC has a 101,115 bps genomic insert that encompasses not only CDKN2A/B, but also the MTAP-009 gene segment, which encodes a 112 amino acid MTAP protein. The detection of losses of this DNA segment in our cases was indicative that both genes could be involved in the malignant transformation of pediatric LGG. By performing expression analysis by qPCR for both the CDKN2A and MTAP genes in cases referred in our series as: LGG 64; 73; 74 and HGG 3; 18; 23, it was evidenced that in the LGG characterized by heterozygous gene losses there was still a “normal” expression of both genes, but in HGG cases, the homozygous loss was associated with a significant down-regulation of both CDKN2A and MTAP genes. The observation that MTAP is also involved in pediatric gliomas is of foremost importance since in very recent surveys [22, 23] it was revealed that MTAP deleted cancers were rendered therapeutically vulnerable at the methionine salvage pathway. Accordingly, to Mavrakis et al. (2016) [22] and Marjon K et al. (2016) [23] deletions at 9p abrogating the MTAP gene lead to the accumulation of its substrate 5′ methilthioadenosine (MTA) and a metabolic “rewiring” of the tumor cells, which rendered them specifically sensible to drugs that inhibit the methyltransferase axis composed by: Methionine Adenosyltransferase II alpha (MAT2A), Protein Arginine Methyltransferase 5 (PMRT5) and Rio domain containing protein 1 (RIOK1).
As aforementioned, HGG18 case of our cohort (Table 1) was diagnosed as a secondary glioblastoma following medullar transplant and radiotherapy of an ALL-B. Reports on the genetic analysis of secondary CNS tumors in children and their previous ALL are exceedingly rare, and the driver versus passenger mutations that underlie leukemia metastasis and the development of secondary tumors are still to be defined [24,25,26]. HGG18 was negative for the presence of BRAF V600E mutation but positive for 9p21 deletion (Table 1). No molecular evaluation for these two genetic alterations could be retrospectively performed in the leukemia cells, but the karyotype observed in bone marrow blasts at relapse revealed a hiperdiploid karyotype, with 54 chromosomes and a gain of two copies of chromosome 9. Although, it could not be ruled out experimentally that in this case a specific CDKN2A/B and MTAP deletion or epigenetic silence of these TSGs were not present ab initio and at relapse in the leukemia cells of the patient, it is possible to consider that in the CNS microenvironment, but not in the medullar microenvironment, CDKN2A/B/MTAP loss and down-regulation contributed to expansion of the patients neoplastic clone, therefore reinforcing the view that chromosomal deletions encompassing these genes are important for glioma progression.
In summary our data allowed us:
-
i)
To verify that the role of BRAF V600E mutation is dependent of other co-occurring genetic alterations, specifically 9p21 deletions that abrogate both the CDKN2A/B and the MTAP tumor suppressor genes;
-
ii)
To demonstrate that 9p21 deletions correlated significantly with poor prognosis indicators and its detection could be used to stratify patients for a stricter surveillance;
-
iii)
To underline the relevance of investigating and defining if glial tumors with CDKN2A/B and MTAP loss may be vulnerable to new forms of therapy, namely those affecting the methionine salvage pathway.
Abbreviations
- ABL1:
-
ABL Proto-Oncogene 1
- BAC:
-
Bacterial Artificial Chromosome
- BRAF:
-
B-RAF Proto-Oncogene, Serine/Threonine Kinase
- CDKN2A :
-
Cyclin Dependent Kinase Inhibitor 2A
- CDKN2B:
-
Cyclin Dependent Kinase Inhibitor 2B
- ERK:
-
Extra Signal-Regulated Kinases
- KLHL9:
-
Kelch Like Family Member 9
- MAPK:
-
Mitogen Activated Protein Kinase
- MTAP :
-
Methylthioadenosine Phosphorilase
- MYB:
-
MYB Proto-Oncogene, Transcription Factor
- MYBL1:
-
MYB Proto-Oncogene Like 1
References
El-Zem R, Bondy M, Wrensch M. Epidemiology of brain tumors. In: Osman FA, editor. Brain Tumors. New Jersey: Humana Press; 2005. p. 3–18.
WHO 2016, Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Ellison DW, Figarella-Branger D, Perry A, Reifenberger G, Deimiling AV. The 2016 WHO classification of tumors of the central nervous system, Revised 4th edition. Lyon: International Agency for Research on Cancer (IARC); 2016.
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumors of the central nervous system, fourth Edi., vol. 114, no. 2. Lyon: International Agency for Research on Cancer (IARC). p. 2007.
Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, Orisme W, Punchihewa C, Parker M, Qaddoumi I, a Boop F, Lu C, Kandoth C, Ding L, Lee R, Huether R, Chen X, Hedlund E, Nagahawatte P, Rusch M, Boggs J, Cheng J, Becksfort J, Ma J, Song G, Li Y, Wei L, Wang J, Shurtleff S, Easton J, Zhao D, Fulton RS, Fulton LL, Dooling DJ, Vadodaria B, Mulder HL, Tang C, Ochoa K, Mullighan CG, Gajjar A, Kriwacki R, Sheer D, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Baker SJ, Ellison DW. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet 2013; 45(6): 602–612.
Ramkissoon LA, Horowitz PM, Craig JM, Ramkissoon SH, Rich BE, Shumacher SE, McKenna A, Lawrence MS, Bergthold G, Brastianos PK, Tabak B, Ducar MD. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. PNAS. 2013;110(20):8188–93.
Jones DTW, Hutter B, Jäger N, Korshunov A, Kool M, Warnatz HJ, Zichner T, Lambert SR, Ryzhova M, Quang DAK, Fontebasso AM, Stütz AM, Hutter S, Zuckermann M, Sturm D, Gronych J, Lasitschka B, Schmidt S, Seker-Cin H, Witt H, Sultan M, Ralser M, Northcott P, Hovestadt V, Bender S, Pfaff E, Stark S, Faury D, Schwartzentruber J, Majewski J, Weber UD, Zapatka M, Raeder B, Schlesner M, Worth CL, Bartholomae CC, von Kalle C, Imbusch CD, Radomski S, Lawerenz C, van Sluis P, Koster J, Volckmann R, Versteeg R, Lehrach H, Monoranu C, Winkler B, Unterberg A, Herold-Mende C, Milde T, Kulozik AE, Ebinger M, Schuhmann MU, Cho YJ, Pomeroy SL, von Deimling A, Witt O, Taylor MD, Wolf S, Karajannis MA, Eberhart CG, Scheurlen W, Hasselblatt M, Ligon KL, Kieran MW, Korbel JO, Yaspo ML, Brors B, Felsberg J, Reifenberger G, Collins VP, Jabado N, Eils R, Lichter P, Pfister SM. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45(8):927–32.
Tatevossian RG, Tatevossian BRG, Tang B, Dalton J, Forshew T, Lawson AR, Ma J, Neale G, Shurtleff SA, Bailey S, Gajjar A, Baker SJ, Sheer D, Ellison DW. MYB upregulation and genetic aberrations in a subset of pediatric low-grade gliomas. Acta Neuropathol. 2010;120(6):731–43.
Gerges N, Fontebasso AM, Albrecht S, Faury D, Jabado N. Pediatric high-grade astrocytomas : a distinct neuro-oncological paradigm. Genome Med. 2013;5(7):66.
Gierke M, Sperveslage J, Schwab D, Beschorner R, Ebinger M, Schuhmann MU, Schittenhelm J. Analysis of IDH1-R132 mutation, BRAF V600 mutation and KIAA1549–BRAF fusion transcript status in central nervous system tumors supports pediatric tumor classification. J Cancer Res Clin Oncol. 2016;142(1):89–100.
Antonelli M, Badiali M, Moi L, Buttarelli FR, Baldi C, Massimino M, Sanson M, Giangaspero F. KIAA1549-BRAF Fusion Gene in Pediatric Brain Tumors of Various Histogenesis. Pediatr Blood Cancer. 2015;62(4):724–7.
Sievert AJ, Lang SS, Boucher KL, Madsen PJ, Slaunwhite E, Choudhari N, Kellet M, Storm PB, Resnick AC. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. PNAS. 2013;110(15):5957–62.
Hawkins C, Walker E, Mohamad N, Zang C, Jacob K, Shirinian M, Alon N, Kahn D, Fried I, Scheinemann K, Tsangaris E, Dirks P, Tressler R, Bouffet E, Jabado N, Tabori U. KIAA1549-BRAF fusion predicts better clinical outcome in paediatric low-grade astrocytoma. Clin Cancer Res. 2011;17(14):4790–8.
Horbinski C, Nikiforova MN, hagenkord JM, Hamilton RL, Pollack IF. Interplay among BRAF, p16, p53, and MIB1 in pediatric low-grade gliomas. Neuro-Oncologia. 2012;14(6):777–89.
Jones DWT, Kocialkowski S, Liu L, Pearson DM, Backlund LM, Ichimura K, Collins VP. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytoma. Cancer Res. 2008;68(21):8673–7.
Schindler G, Capper D, Meyer J, Janzarik W, Omran H, Herold-Mende C, Schmieder K, Wesseling P, Mawrin C, Hasselblatt M, Louis DN, Korshunov A, Pfister SM, Hartmann C, Paulus W, Reifenberger G, Von Deimling A. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):397–405.
Mistry M, Zhukova N, Merico D, Rakopoulos P, Krishnatry R, Shago M, Stavropoulos J, Alon N, Pole JD, Ray PN, Navickiene V, Mangerel J, Remke M, Buczkowicz P, Vamaswamy V, Guerreiro Stucklin A, Li M, Young EJ, Zhang C, Castelo-Branco P, Bakry D, Laughlin S, Shlien A, Chan J, Ligon KL, Rutka JT, Dirks PD, Taylor MD, Greenberg M, Malkin D, Huang A, Bouffet E, Hawkins CE, Tabori U. BRAF Mutation and CDKN2A Deletion Define a Clinically Distinct Subgroup of Childhood Secondary High-Grade Glioma. J Clin Oncol. 2015;33(9):1015–22.
Korshunov A, Ryzhova M, Hovestadt V, Bender S, Sturm D, Capper D, Meyer J, Schrimpf D, Kool M, Northcott P, Zheludkova O, Milde T, Witt O, Kulozik AE, Reifenberger G, Jabado N, Perry A, Lichter P, von Deimling A, Pfister SM, Jones DWT. Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol. 2015;129(5):669–78.
Raabe EH, Lim KS, Kim JM, Meeker A, Mao XG, Nikkhah G, Maciaczyk J, Kahlert U, Jain D, Bar E, Cohen KJ. Eberhart CG. BRAF activation induces transformation and then senescence in human neural stem cells: a pilocytic astrocytoma model. Clin Cancer Res. 2011;17(11):3590–9.
Beillard E, Pallisgaard N, Bi W, van der Velden VHJ, Dee R, van der Schoot CE, Delabesse E, Macintyre E, Gottardi E, Saglio G, Watzinger F, Lion T, van Dongen JJM, Hokland P, Gabert J. Evaluation of candidate control genes for leukemic patients using ‘real-time’ quantitative reverse-transcriptase polimerase chain reaction (RQ-PCR) – a European against Cancer program. Leukemia. 2003;17:2474–86.
Roy DM, Walsh LA, Desrichard A, Huse JT, Wu W, Gao J, Bose P, Lee W, Chan TA. Integrated genomics for pinpointing survival loci within arm-level somatic copy number alterations. Cancer Cell. 2016;29(5):737–50.
Huillard E, Hashizume R, Phillips JJ, Griveau A, Ihrie RA, Aoki Y, Nicolaides T, Perry A, Waldman T, McMahon M, Weiss WA, Petritsch C, James CD, Rowitch DH. Cooperative interactions of BRAFV600E kinase and CDKN2A locus deficiency in pediatric malignant astrocytoma as a basis for rational therapy. PNAS. 2012;109(22):8710–5.
Mavrakis KJ, ER MD 3rd, Mr S, Billy E, Hoffman GR, deWeck A, Ruddy DA, Venkatesan K, Yu J, McAllister G, Stump M, de Beaumont R, Ho S, Yue Y, Liu Y-NY, Yang G, Lin F, Gao H, Kipp DR, Zhao S, Mcnamara JT, Sprague ER, Zheng B, Tallarico J, Mickanin C, Lees E, Pagliarini R, Keen K, Schmelzle T, Hofmann F, Stegmeier F, Sellers WR. Disordered methionie metabolism in MTAP/CDKN2A-deleted cancers to depence of PRMT5. Science. 2016;351(6278):1208–13.
Marjon K, Cameron MJ, Quang P, Clasquin MF, Mandley E, Kunni K, McVay M, Choe S, Kemytsky A, Gross S, Konteatis Z, Murtie J, Blake ML, Travins J, Dorsch M, Biller SA, Marks KM. MTAP deletions in Cancer create vulnerability to targeting of MAT2A/PRMT5/RIOK1 Axis. Cell Rep. 2016;15(3):574–87.
Harrison CJ, Johansson B. Acute Lymphoblastic Leukemia. In: Heim S, Mitelman F, editors. Cancer Cytogenetics. 4th ed: Willey-Blackwell; 2015. p. 233–96.
Alexiou GA. High-grade gliomas in survivors of childhood acute lymphoblastic leukaemia. Childs Nerv Syst. 2009;25(7):779 author reply 781-2.
Alexiou GA, Moschovi M, Georgoulis G, Neroutsou R, Stefanaki K, Sfakianos G, Prodromou N. Anaplastic oligodendrogliomas after treatment of acute lymphoblastic leukemia in children: report of 2 cases. J Neurosurg Pediatr. 2010;5(2):179–83.
Acknowledgements
We thank Paula Gameiro and Helena Alaiz of the Department of Hemato-Oncology of the Portuguese Cancer Institute for the genetic results obtained in the hematologic relapse of case assigned as HGG18 of our series. We thank Sandra Gonçalves for technical assistance, Hélio Belo, Marta Pojo and Paula Gameiro for their invaluable help at the qPCR experiments and to Margarida Morgado for the revision of the English language of the manuscript.
Funding
This study was funded by IPOFG, EPE, Lisbon, Portugal.
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
All authors read and approved the final manuscript. This research has not been and will not be submitted simultaneously to another journal, in whole or in part. The paper reports previously unpublished work. All those named as authors have made a sufficient contribution to the work and have obtained all the necessary consent from their employers or funding bodies. Concept and design: LF, LR, JP, CF, CM. Experiments and procedures: LF, JP, CF, JM, AS, MM, DS, SN, MM, CM, VMN. Data analysis and writing the article: LF, LR.
Corresponding author
Ethics declarations
Competing interest
The authors declare that they have no competing interest.
Ethics approval and consent to participate
The ethical committee of the Portuguese Cancer Institute in Lisbon approved the here in reported (committee’s reference number: UIC/971). Study design and conduction of the study were performed accordingly to high standards of ethics. Written informed consent was obtained prior to any data acquisition, sample collection, or data analysis from the parent or guardian in the case of children under 16.
Consent for publication
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Table S1. Summary of the Clinical and Genetic Data of Glioma Cases. (XLSX 28 kb)
Additional file 2:
Figure S1. Graphical representation of the expression of the CDKN2A and MTAP genes as determined by qPCR in pediatric Low-Grade Gliomas (LGG: 64, 73, 74) and High-Grade Gliomas (HGG: 3, 18, 23 of our series). CN1–3 – non-neoplastic brain lesions used as calibrator controls. (JPG 360 kb)
Additional file 3:
Excel files of TaqMan experiments for the CDKN2A and MTAP genes. (XLSX 127 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Frazão, L., do Carmo Martins, M., Nunes, V.M. et al. BRAF V600E mutation and 9p21: CDKN2A/B and MTAP co-deletions - Markers in the clinical stratification of pediatric gliomas. BMC Cancer 18, 1259 (2018). https://doi.org/10.1186/s12885-018-5120-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-018-5120-0