Abstract
Background
Quantification of acquisition routes of antibiotic resistant bacteria (ARB) is pivotal for understanding transmission dynamics and designing cost-effective interventions. Different methods have been used to quantify the importance of transmission routes, such as relative risks, odds ratios (OR), genomic comparisons and basic reproduction numbers. We systematically reviewed reported estimates on acquisition routes’ contributions of ARB in humans, animals, water and the environment and assessed the methods used to quantify the importance of transmission routes.
Methods
PubMed and EMBASE were searched, resulting in 6054 articles published up until January 1st, 2019. Full text screening was performed on 525 articles and 277 are included.
Results
We extracted 718 estimates with S. aureus (n = 273), E. coli (n = 157) and Enterobacteriaceae (n = 99) being studied most frequently. Most estimates were derived from statistical methods (n = 560), mainly expressed as risks (n = 246) and ORs (n = 239), followed by genetic comparisons (n = 85), modelling (n = 62) and dosage of ARB ingested (n = 17). Transmission routes analysed most frequently were occupational exposure (n = 157), travelling (n = 110) and contacts with carriers (n = 83). Studies were mostly performed in the United States (n = 142), the Netherlands (n = 87) and Germany (n = 60). Comparison of methods was not possible as studies using different methods to estimate the same route were lacking. Due to study heterogeneity not all estimates by the same method could be pooled.
Conclusion
Despite an abundance of published data the relative importance of transmission routes of ARB has not been accurately quantified. Links between exposure and acquisition are often present, but the frequency of exposure is missing, which disables estimation of transmission routes’ importance. To create effective policies reducing ARB, estimates of transmission should be weighed by the frequency of exposure occurrence.
Similar content being viewed by others
Background
Infections caused by antibiotic-resistant bacteria (ARB) are a major global problem. Antibiotic resistance is as ubiquitous as bacteria are, and resistance genes can be found in humans, animals, water and the environment and can be transmitted between and within these reservoirs. The relative attribution of transmission routes differs between bacterial species and resistance elements [1].
While many studies have determined risk factors for acquisition, infection or colonisation with ARB, the global urgency to reduce the prevalence of antibiotic resistance calls for accurate quantification of transmission routes and their relative importance [2]. Chatterjee et al. [2] reviewed studies in which transmission routes were quantified based on odds ratios (OR), and concluded that uniform quantification of relationships between risk factors are required to better understand acquisitions of ARB in humans. Yet the contribution of transmission routes to the total number of acquisitions of ARB bacteria remains unknown. A better understanding of transmission routes and the One Health interplay of ARB is pivotal for developing targeted and cost-effective interventions to effectively reduce ARB transmission.
Accurate quantification of the contribution of different transmission routes to the total burden of ARB firstly requires both the probability to become colonised when exposed as well as the frequency of exposure. For instance, a route with a low acquisition probability per exposure that occurs frequently, may contribute more to the total number of acquisitions than routes with infrequent exposure but with high transmission probability per exposure. The probability to become colonised has been estimated with different methods. Some studies use a risk estimate for colonisation with ARB when exposed and in others the genetic similarity of ARB was derived between source and receiver to establish transmission probability. Another approach is to use mathematical modelling to quantify the basic reproduction number, R0, defined as the expected number of secondary cases caused by a typical infected individual during its period of infectiousness in a completely susceptible population [3]. Such methodological differences may lead to different conclusions on the relative importance of transmission routes. We aimed to systematically review the literature for data on both the probability of acquisition when exposed and frequency of exposure, in order to quantify the importance of different transmission routes of ARB. Yet, despite an abundance of published data, there were no studies providing high-quality data on both parameters. Moreover, heterogeneity in study designs, methods and analytical approaches among studies investigating the same ARB was too high to perform pooled analyses in sensible manner. We, therefore, limit the reporting to a descriptive analysis of the studies included in the systematic review.
Methods
Search strategy
We performed a systematic review in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) protocol [4]. The protocol for this systematic review was registered on PROSPERO (CRD42019136298) and can be accessed at http://www.crd.york.ac.uk/PROSPERO/display_record.php?ID=CRD42019136298 [5].
We identified quantified estimates of transmission routes from humans, animals and environmental reservoirs that resulted in an acquisition of ARB. An acquisition is conceptualized as one or more of: exposure, intake, infection, acquisition, carriage and colonisation. Antibiotic resistance is defined as the resistance of bacteria to one or more antibiotics for which the bacterium is not intrinsically resistant.
PubMed was searched using the search terms shown in Additional file 1: S1 Appendix, resulting in 4576 articles published until January 1st 2019. Secondly, Embase was searched using search terms shown in Additional file 1: S1 Appendix, which resulted in 5195 articles. Articles were not excluded based on publication date, publication type, sample size, significance level or quality Removing duplicates in Endnote and Rayyan resulted in 6054 articles for title/abstract screening. Two researchers separately in- or excluded a random subset of 50 of the 6054 articles, there was an agreement of 100%. Thereafter, one researcher performed the title/abstract screening in Rayyan [6]. In cases of doubt on in- or excluding an abstract/article, the second researcher was consulted. Five-hundred-and-twelve articles were included for full-text screening. After the full-text screening, 277 articles were included. Figure 1 shows a flow diagram of the articles in the review process. The PRISMA 2009 Checklist [4] can be found in Additional file 1: S2 Appendix.
Study selection
Inclusion criteria for the final set of articles were (1) the presence of quantified estimates of (2) a clear acquisition route (3) of ARB. If the study included resistant and non-resistant pathogens the notation “rs” is placed after the transmission estimate to indicate so in the Additional file 1: Table S3.
Studies were excluded if they only reported estimates of the percentage of the source colonised/infected with ARB, for example 10% of the cows on the farm were colonised with MRSA, as this is does not quantify how the cows acquired colonization. We excluded studies on horizontal plasmid transfer, within-host transfer of ARB, for instance from nasal to anal colonization within the same person, and antibiotics as a source of acquisition, because these routes of acquisition do not comprise transmission between different sources. Colonization pressure was excluded because we considered this transmission route too indirect. After applying these inclusion and exclusion criteria, 277 studies and 718 estimates remained.
Data extraction
We extracted the method of estimation, transmission estimate, 95% CI, transmission route, pathogen, whether only resistant pathogens or a pooled estimated for resistant and susceptible pathogens was provided, country where study was performed, size of the study population, author, year of publication, title of manuscript, animal involved in the transmission route and in case of travelling as a transmission route, whether pre-travel screening had been performed. Study characteristics and outcomes were saved in a data extraction table using Microsoft Excel. If estimates based on multivariable analysis were available, these where extracted instead of estimates based on bivariate analysis.
Quality assessment
Only the quality of travel studies was assessed by using a + or − to indicate whether pre-travel screening was performed or not.
Data synthesis and analyses
Reservoirs Twelve reservoirs and cross-reservoirs in which transmission occurs were identified, (1) animal, (2) animal and food, (3) animal and human, (4) animal, human and water, (5) animal and environment, (6) animal and water, (7) human, (8) human and environment, (9) human and food, (10) human and water, (11) environment, and (12) environment and water. Travelling and studies researching an intervention to reduce transmission, e.g., handwashing, were not categorized as a reservoir. Although travelling frequently is considered a “route”, multiple transmission routes, such as food, people and environment, could cause acquisitions during travelling and therefore, we did not appoint travelling to a reservoir.
Transmission routes In all 48 transmission routes were identified and frequencies are shown in the Additional file 1: Table S1.
Methods of estimation Four methods for quantifying transmission routes were identified; [1] based on genetic similarity between ARB or resistance genes in source and receiver; [2] quantifying the dosage of ARB ingested by humans; [3] mathematical modelling to estimate transmission rates; and [4] statistical inference. The latter included calculations of OR, risks, prevalence ratio’s (PR), risk ratio’s (RR) and transmission rates (TR). Methods using genetic similarity estimated percentages of transmissions that had occurred (T, transmission percentage) or estimated a risk being the percentage in which acquisition with a genetically identical strain to the source had occurred from the total sample at risk (R, risk percentage). Modelling methods included R0, attributable percentage of the total number of cases to this route, transmission rates (TRs), cases per day, incidence, incidence rate ratios (IRRs) and acquisition rates (ARs).
Pathogens To estimate transmission routes per ARB, each estimate was categorized per bacterial species if two or more estimates on this species were present, otherwise the estimate was categorized as “other”. The following 19 groups were identified Staphylococcus aureus (S. aureus), Escherichia coli (E. coli), Enterobacteriaceae (estimates pooled for multiple species of Enterobacteriaceae), vancomycin resistant enterococci (VRE), staphylococci, Pseudomonas aeruginosa (P. aeruginosa), Acinetobacter baumannii (A. baumannii), Acinetobacter calcoaceticus (A. calcoaceticus), Campylobacter, Enterococcus faecalis (E. faecalis), Enterococcus faecium (E. faecium), Streptococcus pneumoniae (S. pneumoniae), group B streptococci, salmonella, Staphylococcus epidermidis (S. epidermidis), Staphylococcus pseudintermedius (S. pseudintermedius), Staphylococcus haemolyticus (S. haemolyticus) and other (Enterococci, Mycoplasma hominis, Ureaplasma urealyticum, Klebsiella pneumoniae, Staphylococcus hominis, Staphylococcus cohnii, Klebsiella oxytoca, Haemophilus influenzae, genes of a bacterial species (e.g., blaTEM).
Travelling For travelling countries of destination were grouped in one of the following 11 groups; South Asia (Afghanistan, Bangladesh, Bhutan, Maldives, Nepal, India, Pakistan and Sri Lanka), South East Asia (Indonesia, Cambodia, Laos, Myanmar, Malaysia, Thailand, Vietnam, Australia, Brunei, the Philippines and Singapore), West Asia (Bahrain, Iran, Israel, Jordan, Kuwait, Lebanon, Oman, Qatar, Palestinian territories, Syria, Turkey, Iraq, Saudi Arabia, United Arab Emirates, and Yemen), Central & East Asia (China, Hong Kong, Mongolia, Uzbekistan, Turkmenistan, Tajikistan, Kyrgyzstan, Kazakhstan, East Turkestan, North Korea, South Korea, Japan, and Taiwan), Asia unspecified, Latin America (central Amerika, Mexico and south Amerika), North America, America unspecified, Africa (entire African continent), Europe and multiple regions (multiple of the above regions).
Meta-analysis The initial plan was to perform random effects meta-analyses using the maximum-likelihood estimator per methodology per transmission route per pathogen for studies with the same outcome measure or outcome measures that could be lumped. Yet, as there were not enough studies providing high-quality data on the frequency of exposure and the probability of colonisation, we were unable to quantify the link between probability of colonisation and frequency of exposure. In addition, high heterogeneity in methods, study designs, and analysis performed within categories of ARB transmission routes investigated precluded performance of pooled analyses in sensible manner. We, therefore, limit the reporting to a descriptive analysis of studies included in the systematic review. Our script for the initial meta-analysis in R version 3.6.1. and the datasets generated during the current study are available in the GitHub repository, https://github.com/NoorGo/Transmission_routes_of_antibiotic_resistant_bacteria_systematic_review and as R files and Excel files in Additional file 2, Additional file 3 and Additional file 4.
Results
Study population
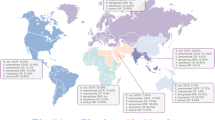
The 277 studies included yielded 718 transmission estimates of 40 transmission routes. Most studies had been performed in the United States (n = 142), the Netherlands (n = 87) and Germany (n = 60) (Fig. 2). Most estimates (n = 560, 78%) were based on statistical inference, predominantly yielding risks (n = 246, 44%) or ORs (n = 239, 43%) (Table 1). Seventeen studies (2%) quantified the dosage of ARB ingested by humans. The most commonly studied transmission routes were occupational exposure (n = 157, 22%), travelling (n = 110, 15%) and contact with carriers (n = 83, 12%) as can be seen in the Additional file 1: Table S1. Most estimates based on travelling (n = 70, 64%) included pre- and post-travel screening. The bacteria most commonly studied were S. aureus (n = 275, 38%), E. coli (n = 160, 22%) and Enterobacteriaceae (n = 121, 14%) as can be seen in Fig. 3 and Additional file 1: Table S2. A list of included articles is reported in the Additional file 1: Table S3. Additional file 1: Table S3 also states which method of estimation was performed, which bacteria and which transmission route was studied for each article.
Cross-reservoir
Cross-reservoir transmission between animals and humans was studied most frequently (n = 211, 29%). Other routes studied were transmission between environments, such as soil, rooms or utensils, and humans (n = 61, 9%), between water and humans (n = 16, 2%), between water and animals (n = 15, 2%), and between animals and the environment (n = 41, 6%) (Fig. 4).
The influence of methodologies on the comparability transmission route importance
Determination of the importance of transmission routes estimated with the above mentioned methodologies was difficult, because not a single transmission route was estimate by all methodologies. This review found that, when applied to the same dataset, risk-based estimates which did not use available genetic data, such as whole-genome sequencing (WGS), tend to overestimate transmission risk compared to estimates including this genetic data as indicated in multiple studies [7,8,9,10,11,12,13,14]. This should be the case, as the genetic data narrow the definition of a potential transmission event, not only should the donor and the recipient have an epidemiological link, both should have the same or a genetically closely related variant of the strain as well.
Importance of transmission routes
Chatterjee et al. [2] provided an overview of ARB transmission to humans estimated by OR, and suggest that uniformly quantifying relationships between risk factors will help to understand the process of ARB acquisition in humans. Here we take all methods to estimate ARB transmission into account, but this still does not allow a proper quantification of the importance of different ARB acquisition routes. We detect a missing link between the risk of acquisition when exposed and frequency of exposure. We aimed to include studies quantifying sources of infections in humans, but only found 1 study which attempted to do so. Cuny et al. [15] take a first step in creating a bigger picture of transmission sources by estimating that, of a large sample of MRSA infections in humans in Germany from 2006 to 2014, 0.14% originated from horses.
We will briefly discuss the transmission routes with the highest acquisition risk given exposure found with the different methods for the largest bacteria groups; S. aureus, E. coli and Enterobacteriaceae.
S. aureus Exposure to pigs and close contact with infected people were frequently identified by multiple methods as high risk transmission routes. For OR, transmission in the human reservoir yielded multiple high estimates, specifically within close contact such as in families. Although OR are high, the absolute risk was estimated low. Based on genetic comparisons contact with a household member yielded high risks for transmission of S. aureus. In other studies, genetical overlap was used to determine the number of acquisitions that could be correctly attributed to the source. Co-occurrence in humans showed the highest proportion of genetically identical strains. When comparing statistically calculated risk estimates to genetical analysis we found that risk estimates of human-to-human transmission based on genetic overlap were the closest to mere statistical risk estimates compared to other transmission routes. For other routes the difference between the estimated risks by these two methods was larger. For example, those thought to be infected by a family member had in 55% [26–80%] an identical strain, which means that in 45% it could not be demonstrated that an infected family member was the source. In this case, a simple risk estimate would have been an overestimation as genetical analysis shows that not colonisations could be attributed to the colonised household member.
Moreover, transmission between the human and animal reservoir, specifically pigs resulted in high OR, usually with large confidence intervals, the highest PR, high risks and the highest risk estimates based on genetic overlap. R0 estimates also indicated pigs as an imported transmitter as pig to pig transmission of S. aureus studied in the Netherlands also had the highest pooled R0 (4.04, 95% CI [2.78–5.86]) [16].
E. coli The human reservoir is a likely route of high E. coli transmission. Close contact with infected people was identified by OR and risk estimates as a high risk E. coli transmission route. Risk estimates based on genetic overlap showed that almost halve of the household members at risk had an E. coli acquisition genetically identical to their household member. OR and risks also indicate animal to environment and animal to human transmission, especially poultry, to be a higher risk route. Travelling to Asia was also identified by OR as high risk.
Enterobacteriaceae Close contact was indicated by OR and PR to be high risk routes. Travelling to Asia was indicated to be high risk by OR and risk estimates. Figures on risk and OR when travelling to regions across the world can be found on GitHub (https://github.com/NoorGo/Transmission_routes_of_antibiotic_resistant_bacteria_systematic_review/tree/Travelling-Figures-ORs-and-Risks).
Discussion
Despite a large number of studies which investigated the probability of acquisition when exposed, we did not identify data that link the probability of acquisition to a frequency of exposure. Information to provide an accurate qualification of the importance of transmission routes was not found.
Nevertheless, some examples allowing quantification do exist. Cuny et al. [15] estimated 0.14% of MRSA infections in humans in Germany between 2006 and 2014 originated from horses. Furthermore, several studies yielded high genetic overlap, OR and PR for bacterial transmission between mother and child during breastfeeding. Yet, as this only occurs in the first years of life this route will not contribute to the total number of transmissions. Regarding the missing link between frequency of exposure and probability of acquisition, multiple studies reported that increased frequency of exposure to animals increased risk of acquisition of MRSA for humans. Increasing hours per day [17, 18], days per week [18, 19], number of animals [19, 20] and number of years of occupational exposure [18, 20] are indicated to increase colonization of farmers which supports our view that a bridge between frequency of exposure and risk of transmission is important.
This systematic review shows that statistical methods, especially ORs and risks, are most often used to estimate transmission routes. Cross-reservoir transmission between humans and animals has been studied most commonly, whereas studies on transmission from the environment are scarce, as also indicated by a previous meta-analysis which only included significant transmission estimates [2].
Comparison of transmission route estimates calculated using different methodologies was difficult as we found few transmission routes that were estimated using multiple methods in comparable populations. Also, no study was identified which conducted all methodologies on the same dataset to study a transmission route. Methods of estimation seems to influence the interpretation of what sources are more likely to cause acquisition. This review found multiple studies indicating that risk-based analyses which do not use genetic data, such as WGS, tend to overestimate the risk compared to studies using genetic data [7,8,9,10,11,12,13,14]. Davis et al. [21] emphasize the added value of whole-genome sequencing, as their previous indicated transmission routes appeared not to have occurred when they compared strains of animals and humans.
Strengths of this review include the assessment and comparison of various methods of estimation, reservoirs and transmission routes so that a complete overview of the current literature on ARB is provided. The first limitation of our study is data collection was done by one reviewer which might have caused discrepancies in data collection and analysis, however measures were taken to limit discrepancies such as a subset which was screened by two authors. Secondly, the extensive literature on the transmission of ARB precludes our search terms from identifying all studies in this area. By including synonyms, we tried to identify as many of the studies as possible. Moreover, publication bias could be present in different degrees for different methods of estimation. For example, whole genome studies that do not identify possible transmission chains may be published less frequently. However, publication bias also occurs for statistical methods of estimation such as OR that do not identify significant routes. Contrary to other reviews [2] we did include non-significant findings, however we cannot control publication bias differences between different methods of estimation.
Conclusion
To conclude, on the one hand, there is a plethora of estimates on the probability of acquisition for ARB transmission routes. On the other hand, the relevance of routes on a population scale is missing due to a missing link between probability of acquisition and frequency of exposure. This lack of knowledge prohibits an accurate prediction of the effectiveness of interventions which target one or more transmission routes. To better quantify acquisition risks in different transmission routes we recommend researchers to perform more focussed and more detailed studies. For instance, population-based studies in well described niches, such as hospitals, other health care facilities, or farms, in which samples can be collected longitudinally in all subjects and environments that may be involved in transmission, as has been performed by Pham et al. [22]. By sampling longitudinally from the environment and subjects one can asses colonization status and the frequency of exposure. Longitudinal studies researching the frequency of exposure, for example how often someone eats raw meat or has contact with animals, and colonization status can provide inside in the link between frequency of exposure and acquisition of ARB. However, measuring the colonization status of the source exposed to, for example is the meat indeed colonised as well, may give problems in practice and therefore, the study design has to be carefully thought out.Large-scale population studies may also reveal reliable information, see for instance Cuny et al. [15] and Mughini-Gras et al. [23]. Yet, such studies will always suffer from missing information as not all subjects and environments can be sampled, requiring statistical adjustments.
Availability of data and materials
The datasets and scripts generated during the current study are available in the GitHub repository, https://github.com/NoorGo/Transmission_routes_of_antibiotic_resistant_bacteria_systematic_review and as R files and Excel files in Additional file 2, Additional file 3 and Additional file 4.
Abbreviations
- A. baumannii :
-
Acinetobacter baumannii
- A. calcoaceticus :
-
Acinetobacter calcoaceticus
- ARB:
-
Antibiotic resistant bacteria
- E. coli :
-
Escherichia coli
- E. faecalis :
-
Enterococcus faecalis
- E. faecium :
-
Enterococcus faecium
- MRSA:
-
Methicillin resistant Staphylococcus aureus
- PRISMA:
-
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- P. aeruginosa :
-
Pseudomonas aeruginosa
- VRE:
-
Vancomycin resistant enterococci
- S. aureus :
-
Staphylococcus aureus
- S. epidermidis :
-
Staphylococcus epidermidis
- S. haemolyticus :
-
Staphylococcus haemolyticus
- S. pneumoniae :
-
Streptococcus pneumoniae
- S. pseudintermedius :
-
Staphylococcus pseudintermedius
- OR:
-
Odds ratios
- WGS:
-
Whole-genome sequencing
References
Holmes AH, Moore LSP, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet (London, England). 2016;387(10014):176–87.
Chatterjee A, Modarai M, Naylor NR, Boyd SE, Atun R, Barlow J, et al. Quantifying drivers of antibiotic resistance in humans: a systematic review. Lancet Infect Dis. 2018;18(12):e368–78.
Diekmann O, Heesterbeek JA, Metz JA. On the definition and the computation of the basic reproduction ratio R0 in models for infectious diseases in heterogeneous populations. J Math Biol. 1990;28(4):365–82.
Moher D, Liberati A, Tetzlaff J, Altman DG, Garoup TP. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLOS Med [Internet]. 2009;6(7):e1000097. https://doi.org/10.1371/journal.pmed.1000097.
Godijk NG, Bootsma MCJ. What is the estimated importance of transmission routes for antimicrobial-resistant bacteria according to different methodologies? PROSPERO 2019 CRD42019136298 [Internet]. Available from: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42019136298.
Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan–-a web and mobile app for systematic reviews. Syst Rev [Internet]. 2016;5(1):210. https://doi.org/10.1186/s13643-016-0384-4.
Tandé D, Boisramé-Gastrin S, Münck MR, Héry-Arnaud G, Gouriou S, Jallot N, et al. Intrafamilial transmission of extended-spectrum-β-lactamase-producing Escherichia coli and Salmonella enterica Babelsberg among the families of internationally adopted children. J Antimicrob Chemother [Internet]. 2010;65(5):859–65. https://doi.org/10.1093/jac/dkq068.
Mutters NT, Brooke RJ, Frank U, Heeg K. Low risk of apparent transmission of vancomycin-resistant Enterococci from bacteraemic patients to hospitalized contacts. Am J Infect Control. 2013;41(9):778–81.
De Martino L, Lucido M, Mallardo K, Facello B, Mallardo M, Iovane G, et al. Methicillin-resistant staphylococci isolated from healthy horses and horse personnel in Italy. J Vet Diagn Invest. 2010;22(1):77–82.
Gordoncillo MJ, Abdujamilova N, Perri M, Donabedian S, Zervos M, Bartlett PC. Detection of methicillin-resistant Staphylococcus aureus (MRSA) in backyard pigs and their owners, Michigan, USA. Zoonoses Public Health. 2012;59(3):212–6.
Knetsch CW, Connor TR, Mutreja A, van Dorp SM, Sanders IM, Browne HP, et al. Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011. Euro Surveill. 2014;19(45):20954.
Nerby JM, Gorwitz R, Lesher L, Juni B, Jawahir S, Lynfield R, et al. Risk factors for household transmission of community-associated methicillin-resistant Staphylococcus aureus. Pediatr Infect Dis J. 2011;30(11):927–32.
McCallum SJ, Gallagher MJ, Corkill JE, Hart CA, Ledson MJ, Walshaw MJ. Spread of an epidemic Pseudomonas aeruginosa strain from a patient with cystic fibrosis (CF) to non-CF relatives. Thorax. 2002;57(6):559–60.
Ozaki K, Takano M, Higuchi W, Takano T, Yabe S, Nitahara Y, et al. Genotypes, intrafamilial transmission, and virulence potential of nasal methicillin-resistant Staphylococcus aureus from children in the community. J Infect Chemother. 2009;15(2):84–91.
Cuny C, Abdelbary M, Koeck R, Layer F, Werner G, Witte W. Methicillin resistant Staphylococcus aureus from infections in horses in Germany: significance for infections in humans. Int J Med Microbiol [Internet]. 2015;305:172. https://doi.org/10.1016/j.onehlt.2015.11.004.
Broens EM, Graat EAM, van de Giessen AW, Broekhuizen-Stins MJ, de Jong MCM. Quantification of transmission of livestock-associated methicillin resistant Staphylococcus aureus in pigs. Vet Microbiol. 2012;155(2–4):381–8.
Dohmen W, Bonten MJM, Bos MEH, van Marm S, Scharringa J, Wagenaar JA, et al. Carriage of extended-spectrum beta-lactamases in pig farmers is associated with occurrence in pigs. Clin Microbiol Infect. 2015;21(10):917–23.
Li L, Chen Z, Guo D, Li S, Huang J, Wang X, et al. Nasal carriage of methicillin-resistant coagulase-negative staphylococci in healthy humans is associated with occupational pig contact in a dose-response manner. Vet Microbiol. 2017;208:231–8.
Mascaro V, Leonetti M, Nobile CGA, Barbadoro P, Ponzio E, Recanatini C, et al. Prevalence of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) among farm and slaughterhouse workers in Italy. J Occup Environ Med. 2018;60(8):E416–25.
Rasamiravaka T, Andriatsitohanana TT, Rasamindrakotroka A. Evaluation of methicillin-resistant Staphylococcus aureus nasal carriage in malagasy pig and poultry non-industrial farmers. J Infect Dev Ctries. 2017;11(2):129–35.
Davis MF, Misic AM, Morris DO, Moss JT, Tolomeo P, Beiting DP, et al. Genome sequencing reveals strain dynamics of methicillin-resistant Staphylococcus aureus in the same household in the context of clinical disease in a person and a dog. Vet Microbiol. 2015;180(3–4):304–7.
Pham TM, Kretzschmar M, Bertrand X, Bootsma M, Consortium on behalf of C-M. Tracking Pseudomonas aeruginosa transmissions due to environmental contamination after discharge in ICUs using mathematical models. PLOS Comput Biol [Internet]. 2019;15(8):e1006697. https://doi.org/10.1371/journal.pcbi.1006697.
Mughini-Gras L, Dorado-García A, van Duijkeren E, van den Bunt G, Dierikx CM, Bonten MJM, et al. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: a population-based modelling study. Lancet Planet Heal. 2019;3(8):e357–69.
Rosen K, Roesler U, Merle R, Friese A. Persistent and transient airborne MRSA colonization of piglets in a newly established animal model. Front Microbiol. 2018;9(JUL):1542.
Dierikx CM, van der Goot J, van Essen-Zandbergen A, Mevius DJ. Dynamics of cefotaxime resistant Escherichia coli in broilers in the first week of life. Vet Microbiol. 2018;222:64–8.
van Duijkeren E, Kamphuis M, van der Mije IC, Laarhoven LM, Duim B, Wagenaar JA, et al. Transmission of methicillin-resistant Staphylococcus pseudintermedius between infected dogs and cats and contact pets, humans and the environment in households and veterinary clinics. Vet Microbiol. 2011;150(3–4):338–43.
Loeffler A, Pfeiffer DU, Lindsay JA, Soares-Magalhaes R, Lloyd DH. Lack of transmission of methicillin-resistant Staphylococcus aureus (MRSA) between apparently healthy dogs in a rescue kennel. Vet Microbiol. 2010;141(1–2):178–81.
Manga I, Hasman H, Smidkova J, Medvecky M, Dolejska M, Cizek A. Fecal carriage and whole-genome sequencing-assisted characterization of CMY-2 beta-lactamase-producing Escherichia coli in calves at Czech dairy cow farm. Foodborne Pathog Dis. 2019;16(1):42–53.
Huijbers PMC, Graat EAM, van Hoek AHAM, Veenman C, de Jong MCM, van Duijkeren E. Transmission dynamics of extended-spectrum β-lactamase and AmpC β-lactamase-producing Escherichia coli in a broiler flock without antibiotic use. Prev Vet Med. 2016;131:12–9.
Riccobono E, Pallecchi L, Mantella A, Bartalesi F, Zeballos IC, Trigoso C, et al. Carriage of antibiotic-resistant Escherichia coli among healthy children and home-raised chickens: a household study in a resource-limited setting. Microb Drug Resist. 2012;18(1):83–7.
Weese JS, Dick H, Willey BM, McGeer A, Kreiswirth BN, Innis B, et al. Suspected transmission of methicillin-resistant Staphylococcus aureus between domestic pets and humans in veterinary clinics and in the household. Vet Microbiol. 2006;115(1–3):148–55.
Windahl U, Ågren J, Holst BS, Börjesson S. Colonization with methicillin-resistant Staphylococcus pseudintermedius in multi-dog households: a longitudinal study using whole genome sequencing. Vet Microbiol. 2016;189:8–14.
Graveland H, Wagenaar JA, Verstappen KMHW, Oosting-van Schothorst I, Heederik DJJ, Bos MEH. Dynamics of MRSA carriage in veal calves: a longitudinal field study. Prev Vet Med. 2012;107(3–4):180–6.
Gao LL, Tan Y, Zhang XD, Hu JQ, Miao ZM, Wei LM, et al. Emissions of Escherichia coli carrying extended-spectrum β-lactamase resistance from pig farms to the surrounding environment. Int J Environ Res Public Health. 2015;12(4):4203–13.
Laube H, Friese A, von Salviati C, Guerra B, Rösler U. Transmission of ESBL/AmpC-producing Escherichia coli from broiler chicken farms to surrounding areas. Vet Microbiol. 2014;172(3–4):519–27.
Moodley A, Guardabassi L. Transmission of IncN plasmids carrying blaCTX-M-1 between commensal Escherichia coli in pigs and farm workers. Antimicrob Agents Chemother. 2009;53(4):1709–11.
Gao L, Hu J, Zhang X, Wei L, Li S, Miao Z, et al. Application of swine manure on agricultural fields contributes to extended- spectrum ß-lactamase producing Escherichia coli spread in Tai’an, China. Front Microbiol. 2015;6(MAR):313.
Leite-Martins L, Meireles D, Bessa LJ, Mendes Â, De Matos AJ, Martins Da Costa P. Spread of multidrug-resistant Enterococcus faecalis within the household setting. Microb Drug Resist. 2014;20(5):501–7.
Schmithausen RM, Schulze-Geisthoevel SV, Stemmer F, El-Jade M, Reif M, Hack S, et al. Analysis of transmission of MRSA and ESBL-E among pigs and farm personnel. PLoS ONE. 2015;10(9):e0138173.
Davis MF, Pisanic N, Rhodes SM, Brown A, Keller H, Nadimpalli M, et al. Occurrence of Staphylococcus aureus in swine and swine workplace environments on industrial and antibiotic-free hog operations in North Carolina, USA: a One Health pilot study. Environ Res. 2018;163:88–96.
Fu J, Yang D, Jin M, Liu W, Zhao X, Li C, et al. Aquatic animals promote antibiotic resistance gene dissemination in water via conjugation: role of different regions within the zebra fish intestinal tract, and impact on fish intestinal microbiota. Mol Ecol. 2017;26(19):5318–33.
Parm U, Metsvaht T, Sepp E, Ilmoja ML, Pisarev H, Pauskar M, et al. Risk factors associated with gut and nasopharyngeal colonization by common Gram-negative species and yeasts in neonatal intensive care units patients. Early Hum Dev. 2011;87(6):391–9.
Chen CJ, Hsu KH, Lin TY, Hwang KP, Chen PY, Huang YC. Factors associated with nasal colonization of methicillin-resistant Staphylococcus aureus among healthy children in Taiwan. J Clin Microbiol. 2011;49(1):131–7.
Isendahl J, Turlej-Rogacka A, Manjuba C, Rodrigues A, Giske CG, Nauclér P. Fecal carriage of ESBL-producing E. coli and K. pneumoniae in children in Guinea-Bissau: a hospital-based cross-sectional study. PLoS ONE. 2012;7(12):e51981.
Hijazi SM, Fawzi MA, Ali FM, Abd El Galil KH. Multidrug-resistant ESBL-producing Enterobacteriaceae and associated risk factors in community infants in Lebanon. J Infect Dev Ctries. 2016;10(9):947–55.
Koliou MG, Andreou K, Lamnisos D, Lavranos G, Iakovides P, Economou C, et al. Risk factors for carriage of Streptococcus pneumoniae in children. BMC Pediatr. 2018;18(1):144.
Nakamura K, Kaneko M, Abe Y, Yamamoto N, Mori H, Yoshida A, et al. Outbreak of extended-spectrum beta-lactamase-producing Escherichia coli transmitted through breast milk sharing in a neonatal intensive care unit. J Hosp Infect. 2016;92(1):42–6.
Nordberg V, Quizhpe Peralta A, Galindo T, Turlej-Rogacka A, Iversen A, Giske CG, et al. High proportion of intestinal colonization with successful epidemic clones of esbl-producing enterobacteriaceae in a neonatal intensive care unit in Ecuador. PLoS ONE. 2013;8(10):e76597.
Çftç E, Aysev ÜDD, Ïnce E, Gürz H. Investigation of risk factors for penicillin-resistant Streptococcus pneumoniae carriage in Turkish children. Pediatr Int. 2001;43(4):385–90.
Gastelum DT, Dassey D, Mascola L, Yasuda LM. Transmission of community-associated methicillin-resistant Staphylococcus aureus from breast milk in the neonatal intensive care unit. Pediatr Infect Dis J. 2005;24(12):1122–4.
Benito D, Lozano C, Jiménez E, Albújar M, Gómez A, Rodríguez JM, et al. Characterization of Staphylococcus aureus strains isolated from faeces of healthy neonates and potential mother-to-infant microbial transmission through breastfeeding. FEMS Microbiol Ecol. 2015;91(3).
Gueimonde M, Salminen S, Isolauri E. Presence of specific antibiotic (tet) resistance genes in infant faecal microbiota. FEMS Immunol Med Microbiol. 2006;48(1):21–5.
Jackson SS, Harris AD, Magder LS, Stafford KA, Johnson JK, Miller LG, et al. Bacterial burden is associated with increased transmission to health care workers from patients colonized with vancomycin-resistant Enterococcus. Am J Infect Control. 2019;47(1):13–7.
Morgan DJ, Liang SY, Smith CL, Johnson JK, Harris AD, Furuno JP, et al. Frequent multidrug-resistant Acinetobacter baumannii contamination of gloves, gowns, and hands of healthcare workers. Infect Control Hosp Epidemiol. 2010;31(7):716–21.
Schwartz-Neiderman A, Braun T, Fallach N, Schwartz D, Carmeli Y, Schechner V. Risk factors for carbapenemase-producing carbapenem-resistant enterobacteriaceae (CP-CRE) acquisition among contacts of newly diagnosed CP-CRE patients. Infect Control Hosp Epidemiol. 2016;37(10):1219–25.
Grabsch EA, Burrell LJ, Padiglione A, O’Keeffe JM, Ballard S, Grayson ML. Risk of environmental and healthcare worker contamination with vancomycin-resistant enterococci during outpatient procedures and hemodialysis. Infect Control Hosp Epidemiol. 2006;27(3):287–93.
Snyder GM, Thorn KA, Furuno JP, Perencevich EN, Roghmann M-C, Strauss SM, et al. Detection of methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci on the gowns and gloves of healthcare workers. Infect Control Hosp Epidemiol. 2008;29(7):583–9.
El Shafie SS, Alishaq M, Leni GM. Investigation of an outbreak of multidrug-resistant Acinetobacter baumannii in trauma intensive care unit. J Hosp Infect. 2004;56(2):101–5.
Senn L, Clerc O, Zanetti G, Basset P, Prodhom G, Gordon NC, et al. The stealthy superbug: the role of asymptomatic enteric carriage in maintaining a long-term hospital outbreak of ST228 methicillin-resistant Staphylococcus aureus. MBio. 2016;7(1):e02039-e2115.
Rydberg J, Cederberg Å. Intrafamilial spreading of Escherichia coli resistant to trimethoprim. Scand J Infect Dis. 1986;18(5):457–60.
Frénay HME, Vandenbroucke-Grauls CMJE, Molkenboer MJCH, Verhoef J. Long-term carriage, and transmission of methicillin-resistant Staphylococcus aureus after discharge from hospital. J Hosp Infect. 1992;22(3):207–15.
McBryde ES, Bradley LC, Whitby M, McElwain DLS. An investigation of contact transmission of methicillin-resistant Staphylococcus aureus. J Hosp Infect. 2004;58(2):104–8.
Schaumburg F, Alabi AS, Mombo-Ngoma G, Kaba H, Zoleko RM, Diop DA, et al. Transmission of Staphylococcus aureus between mothers and infants in an African setting. Clin Microbiol Infect. 2014;20(6):O390–6.
Torres E, López-Cerero L, Morales I, Navarro MD, Rodríguez-Baño J, Pascual A. Prevalence and transmission dynamics of Escherichia coli ST131 among contacts of infected community and hospitalized patients. Clin Microbiol Infect. 2018;24(6):618–23.
Zhou Q, Moore C, Eden S, Tong A, McGeer A. Factors associated with acquisition of vancomycin-resistant enterococci (VRE) in roommate contacts of patients colonized or infected with VRE in a tertiary care hospital. Infect Control Hosp Epidemiol. 2008;29(5):398–403.
Hedin G, Hambraeus A. Multiply antibiotic-resistant Staphylococcus epidermidis in patients, staff and environment–a one-week survey in a bone marrow transplant unit. J Hosp Infect. 1991;17(2):95–106.
Pierce R, Lessler J, Popoola VO, Milstone AM. Meticillin-resistant Staphylococcus aureus (MRSA) acquisition risk in an endemic neonatal intensive care unit with an active surveillance culture and decolonization programme. J Hosp Infect. 2017;95(1):91–7.
Calfee DP, Durbin LJ, Germanson TP, Toney DM, Smith EB, Farr BM. Spread of methicillin-resistant Staphylococcus aureus (MRSA) among household contacts of individuals with nosocomially acquired MRSA. Infect Control Hosp Epidemiol. 2003;24(6):422–6.
Domenech de Celles M, Salomon J, Marinier A, Lawrence C, Gaillard J-L, Herrmann J-L, et al. Identifying more epidemic clones during a hospital outbreak of multidrug-resistant Acinetobacter baumannii. PLoS ONE. 2012;7(9):e45758.
Christopher S, Verghis RM, Antonisamy B, Sowmyanarayanan TV, Brahmadathan KN, Kang G, et al. Transmission dynamics of methicillin-resistant Staphylococcus aureus in a medical intensive care unit in India. PLoS ONE. 2011;6(7):e20604.
Austin DJ, Bonten MJ, Weinstein RA, Slaughter S, Anderson RM. Vancomycin-resistant enterococci in intensive-care hospital settings: transmission dynamics, persistence, and the impact of infection control programs. Proc Natl Acad Sci U S A. 1999;96(12):6908–13.
Plipat N, Spicknall IH, Koopman JS, Eisenberg JNS. The dynamics of methicillin-resistant Staphylococcus aureus exposure in a hospital model and the potential for environmental intervention. BMC Infect Dis. 2013;13(1):595.
Mikolajczyk RT, Kauermann G, Sagel U, Kretzschmar M. Mixture model to assess the extent of cross-transmission of multidrug-resistant pathogens in hospitals. Infect Control Hosp Epidemiol. 2009;30(8):730–6.
Cheah ALY, Cheng AC, Spelman D, Nation RL, Kong DCM, McBryde ES. Mathematical modelling of vancomycin-resistant enterococci transmission during passive surveillance and active surveillance with contact isolation highlights the need to identify and address the source of acquisition. BMC Infect Dis. 2018;18(1):511.
Cooper BS, Kypraios T, Batra R, Wyncoll D, Tosas O, Edgeworth JD. Quantifying type-specific reproduction numbers for nosocomial pathogens: evidence for heightened transmission of an Asian sequence type 239 MRSA clone. PLoS Comput Biol. 2012;8(4):e1002454.
Domenech de Celles M, Opatowski L, Salomon J, Varon E, Carbon C, Boelle P-Y, et al. Intrinsic epidemicity of Streptococcus pneumoniae depends on strain serotype and antibiotic susceptibility pattern. Antimicrob Agents Chemother. 2011;55(11):5255–61.
Forrester M, Pettitt AN. Use of stochastic epidemic modeling to quantify transmission rates of colonization with methicillin-resistant Staphylococcus aureus in an intensive care unit. Infect Control Hosp Epidemiol. 2005;26(7):598–606.
Pettitt AN, Forrester ML, Gibson GJ. Bayesian inference of hospital-acquired infectious diseases and control measures given imperfect surveillance data. Biostatistics. 2007;8(2):383–401.
Hetem DJ, Westh H, Boye K, Jarløv JO, Bonten MJM, Bootsma MCJ. Nosocomial transmission of community-associated methicillin-resistant Staphylococcus aureus in Danish hospitals. J Antimicrob Chemother. 2012;67(7):1775–80.
Mikolajczyk RT, Sagel U, Bornemann R, Krämer A, Kretzschmar M. A statistical method for estimating the proportion of cases resulting from cross-transmission of multi-resistant pathogens in an intensive care unit. J Hosp Infect. 2007;65(2):149–55.
Simon CP, Percha B, Riolo R, Foxman B. Modeling bacterial colonization and infection routes in health care settings: analytic and numerical approaches. J Theor Biol. 2013;334:187–99.
López-García M, Kypraios T. A unified stochastic modelling framework for the spread of nosocomial infections. J R Soc Interface. 2018;15(143).
McBryde ES, Pettitt AN, McElwain DLS. A stochastic mathematical model of methicillin resistant Staphylococcus aureus transmission in an intensive care unit: predicting the impact of interventions. J Theor Biol. 2007;245(3):470–81.
Pelupessy I, Bonten MJM, Diekmann O. How to assess the relative importance of different colonization routes of pathogens within hospital settings. Proc Natl Acad Sci U S A. 2002;99(8):5601–5.
Wang L, Ruan S. Modeling nosocomial infections of methicillin-resistant Staphylococcus aureus with environment contamination. Sci Rep. 2017;7(1):580.
Lo W-U, Ho P-L, Chow K-H, Lai EL, Yeung F, Chiu SS. Fecal carriage of CTXM type extended-spectrum beta-lactamase-producing organisms by children and their household contacts. J Infect. 2010;60(4):286–92.
Givon-Lavi N, Fraser D, Porat N, Dagan R. Spread of Streptococcus pneumoniae and antibiotic-resistant S. pneumoniae from day-care center attendees to their younger siblings. J Infect Dis. 2002;186(11):1608–14.
Alam MT, Read TD, Petit RA, Boyle-Vavra S, Miller LG, Eells SJ, et al. Transmission and microevolution of USA300 MRSA in U.S. households: evidence from whole-genome sequencing. MBio. 2015;6(2):e00054.
Alves M, Lemire A, Decré D, Margetis D, Bigé N, Pichereau C, et al. Extended-spectrum beta-lactamase - producing enterobacteriaceae in the intensive care unit: acquisition does not mean cross-transmission. BMC Infect Dis. 2016;16(1):147.
Chidekel AS, Steele-Moore L, Padman R, Pellegrino V, Berg D, Klein J. Nasopharyngeal colonization in children with cystic fibrosis: antibiotic resistance and intrafamilial spread. Infect Dis Clin Pract [Internet]. 2000;9(8):333–6.
Cochard H, Aubier B, Quentin R, van der Mee-Marquet N. Extended-spectrum β -lactamase–producing enterobacteriaceae in french nursing homes: an association between high carriage rate among residents, environmental contamination, poor conformity with good hygiene practice, and putative resident-to-resident tra. Infect Control Hosp Epidemiol. 2014;35(4):384–9.
Crombach WHJ, Dijkshoorn L, van Noort-Klaassen M, Niessen J, van Knippenberg-Gordebeke G. Control of an epidemic spread of a multi-resistant strain of Acinetobacter calcoaceticus in a hospital. Intensive Care Med. 1989;15(3):166–70.
Cuny C, Nathaus R, Layer F, Strommenger B, Altmann D, Witte W. Nasal colonization of humans with methicillin-resistant Staphylococcus aureus (MRSA) CC398 with and without exposure to pigs. PLoS One [Internet]. 2009;4(8):e6800–e6800. https://doi.org/10.1371/journal.pone.0006800.
Eveillard M, Martin Y, Hidri N, Boussougant Y, Joly-Guillou M-L. Carriage of methicillin-resistant Staphylococcus aureus among hospital employees: prevalence, duration, and transmission to households. Infect Control Hosp Epidemiol. 2004;25(2):114–20.
Griffith SJ, Nathan C, Selander RK, Chamberlin W, Gordon S, Kabins S, et al. The epidemiology of Pseudomonas aeruginosa in oncology patients in a general hospital. J Infect Dis. 1989;160(6):1030–6.
Gustafsson EB, Ringberg H, Johansson PJH. MRSA in children from foreign countries adopted to Swedish families. Acta Paediatr Int J Paediatr. 2007;96(1):105–8.
Normark BH, Christensson B, Sandgren A, Noreen B, Sylvan S, Burman LG, et al. Clonal analysis of Streptococcus pneumoniae nonsusceptible to penicillin at day-care centers with index cases, in a region with low incidence of resistance: emergence of an invasive type 35B clone among carriers. Microb Drug Resist. 2003;9(4):337–44.
Johansson PJH, Gustafsson EB, Ringberg H. High prevalence of MRSA in household contacts. Scand J Infect Dis. 2007;39(9):764–8.
Johnson JK, Smith G, Lee MS, Venezia RA, Stine OC, Nataro JP, et al. The role of patient-to-patient transmission in the acquisition of imipenem-resistant Pseudomonas aeruginosa colonization in the intensive care unit. J Infect Dis. 2009;200(6):900–5.
Kojima Y, Harada S, Aoki K, Ishii Y, Sawa T, Hasegawa K, et al. Spread of CTX-M-15 extended-spectrum β-lactamase-producing Escherichia coli isolates through household contact and plasmid transfer. J Clin Microbiol [Internet]. 2014;52(5):1783–5. https://doi.org/10.1128/JCM.03342-13.
Maataoui N, Mayet A, Duron S, Delacour H, Mentré F, Laouenan C, et al. High acquisition rate of extended-spectrum β-lactamase-producing Enterobacteriaceae among French military personnel on mission abroad, without evidence of inter-individual transmission. Clin Microbiol Infect. 2019;25(5):631.e1-631.e9.
Mitsuda T, Arai K, Ibe M, Imagawa T, Tomono N, Yokota S. The influence of methicillin-resistant Staphylococcus aureus (MRSA) carriers in a nursery and transmission of MRSA to their households. J Hosp Infect. 1999;42(1):45–51.
Cooper BS, Medley GF, Stone SP, Kibbler CC, Cookson BD, Roberts JA, et al. Methicillin-resistant Staphylococcus aureus in hospitals and the community: stealth dynamics and control catastrophes. Proc Natl Acad Sci U S A. 2004;101(27):10223–8.
Toleman MS, Reuter S, Coll F, Harrison EM, Blane B, Brown NM, et al. Systematic surveillance detects multiple silent introductions and household transmission of methicillin-resistant Staphylococcus aureus USA300 in the East of England. J Infect Dis. 2016;214(3):447–53.
Kariuki S, Revathi G, Kariuki N, Kiiru J, Mwituria J, Muyodi J, et al. Invasive multidrug-resistant non-typhoidal Salmonella infections in Africa: zoonotic or anthroponotic transmission? J Med Microbiol. 2006;55(5):585–91.
Geffers C, Farr BM. Risk of transmission of nosocomial methicillin-resistant Staphylococcus aureus (MRSA) from patients colonized with MRSA. Vol. 26, Infection control and hospital epidemiology. United States; 2005. p. 114–5.
Martinez JA, Ruthazer R, Hansjosten K, Barefoot L, Snydman DR. Role of environmental contamination as a risk factor for acquisition of vancomycin-resistant enterococci in patients treated in a medical intensive care unit. Arch Intern Med. 2003;163(16):1905–12.
Salm F, Deja M, Gastmeier P, Kola A, Hansen S, Behnke M, et al. Prolonged outbreak of clonal MDR Pseudomonas aeruginosa on an intensive care unit: contaminated sinks and contamination of ultra-filtrate bags as possible route of transmission? Antimicrob Resist Infect Control. 2016;5(1):53.
Leistner R, Meyer E, Gastmeier P, Pfeifer Y, Eller C, Dem P, et al. Risk factors associated with the community-acquired colonization of extended-spectrum beta-lactamase (ESBL) positive Escherichia coli an exploratory case-control study. PLoS ONE. 2013;8(9):e74323.
Lietzau S, Raum E, von Baum H, Marre R, Brenner H. Clustering of antibiotic resistance of E. coli in couples: suggestion for a major role of conjugal transmission. BMC Infect Dis. 2006;6:119.
Nadimpalli M, Vuthy Y, de Lauzanne A, Fabre L, Criscuolo A, Gouali M, et al. Meat and fish as sources of extended-spectrum beta-lactamase-producing Escherichia coli, Cambodia. Emerg Infect Dis. 2019;25(1).
Samore MH, Tonnerre C, Hannah EL, Stoddard GJ, Borotkanics RJ, Haddadin B, et al. Impact of outpatient antibiotic use on carriage of ampicillin-resistant Escherichia coli. Antimicrob Agents Chemother. 2011;55(3):1135–41.
Börjesson S, Ny S, Egervärn M, Bergström J, Rosengren Å, Englund S, et al. Limited dissemination of extended-spectrum β-lactamase–and plasmid-encoded AmpC–producing Escherichia coli from food and farm animals, Sweden. Emerg Infect Dis. 2016;22(4):634–40.
Sorensen TL, Blom M, Monnet DL, Frimodt-Moller N, Poulsen RL, Espersen F. Transient intestinal carriage after ingestion of antibiotic-resistant Enterococcus faecium from chicken and pork. N Engl J Med. 2001;345(16):1161–6.
Duckro AN, Blom DW, Lyle EA, Weinstein RA, Hayden MK. Transfer of vancomycin-resistant enterococci via health care worker hands. Arch Intern Med. 2005;165(3):302–7.
Kotay S, Chai W, Guilford W, Barry K, Mathers AJ. Spread from the sink to the patient: In situ study using green fluorescent protein (GFP)-expressing Escherichia coli to model bacterial dispersion from hand-washing sink-trap reservoirs. Appl Environ Microbiol. 2017;83(8).
Habteselassie MY, Bischoff M, Applegate B, Reuhs B, Turco RF. Understanding the role of agricultural practices in the potential colonization and contamination by Escherichia coli in the rhizospheres of fresh produce. J Food Prot. 2010;73(11):2001–9.
Ferreira JP, Anderson KL, Correa MT, Lyman R, Ruffin F, Reller LB, et al. Transmission of mrsa between companion animals and infected human patients presenting to outpatient medical care facilities. PLoS ONE. 2011;6(11):e26978.
Fornasini M, Reves RR, Murray BE, Morrow AL, Pickering LK. Trimethoprim-resistant Escherichia coli in households of children attending day care centers. J Infect Dis. 1992;166(2):326–30.
Samore MH, Magill MK, Alder SC, Severina E, Morrison-De Boer L, Lyon JL, et al. High rates of multiple antibiotic resistance in Streptococcus pneumoniae from healthy children living in isolated rural communities: association with cephalosporin use and intrafamilial transmission. Pediatrics. 2001;108(4):856–65.
Cluzet VC, Gerber JS, Nachamkin I, Metlay JP, Zaoutis TE, Davis MF, et al. Risk factors for recurrent colonization with methicillin-resistant Staphylococcus aureus in community-dwelling adults and children. Infect Control Hosp Epidemiol. 2015;36(7):786–93.
Stewardson AJ, Vervoort J, Adriaenssens N, Coenen S, Godycki-Cwirko M, Kowalczyk A, et al. Effect of outpatient antibiotics for urinary tract infections on antimicrobial resistance among commensal Enterobacteriaceae: a multinational prospective cohort study. Clin Microbiol Infect. 2018;24(9):972–9.
Paltansing S, Vlot JA, Kraakman MEM, Mesman R, Bruijning ML, Bernards AT, et al. Extended-spectrum beta-lactamase-producing enterobacteriaceae among travelers from the Netherlands. Emerg Infect Dis. 2013;19(8):1206–13.
Zetsma JW, Spanjaard L, Dankert J. Carrier state and spread of methicillin resistant Staphylococcus aureus following hospital discharge. Ned Tijdschr Geneeskd. 1993;137(47):2428–31.
Garcia-Graells C, Antoine J, Larsen J, Catry B, Skov R, Denis O. Livestock veterinarians at high risk of acquiring methicillin-resistant Staphylococcus aureus ST398. Epidemiol Infect. 2012;140(3):383–9.
Lo W-T, Lin W-J, Tseng M-H, Wang S-R, Wang C-C. Dissemination of Methicillin-resistant Staphylococcus aureus among Healthy Children in Northern Taiwan. J Med Sci. 2010;1:30.
Lu PL, Tsai JC, Chiu YW, Chang FY, Chen YW, Hsiao CF, et al. Methicillin-resistant Staphylococcus aureus carriage, infection and transmission in dialysis patients, healthcare workers and their family members. Nephrol Dial Transplant. 2008;23(5):1659–65.
Neyra RC, Frisancho JA, Rinsky JL, Resnick C, Carroll KC, Rule AM, et al. Multidrug-resistant and methicillin-resistant Staphylococcus aureus (MRSA) in hog slaughter and processing plant workers and their community in North Carolina (USA). Environ Health Perspect. 2014;122(5):471–7.
Würtz ET, Bønløkke JH, Urth TR, Larsen J, Islam MZ, Sigsgaard T, et al. No apparent transmission of livestock-associated methicillin-resistant Staphylococcus aureus CC398 in a survey of staff at a regional Danish hospital. Antimicrob Resist Infect Control. 2017;6(1):126.
Baran JJ, Ramanathan J, Riederer KM, Khatib R. Stool colonization with vancomycin-resistant enterococci in healthcare workers and their households. Infect Control Hosp Epidemiol. 2002;23(1):23–6.
Maree CL, Eells SJ, Tan J, Bancroft EA, Malek M, Harawa NT, et al. Risk factors for infection and colonization with community-associated methicillin-resistant Staphylococcus aureus in the Los Angeles County jail: a case-control study. Clin Infect Dis. 2010;51(11):1248–57.
Nilsson P, Laurell MH. Carriage of penicillin-resistant Streptococcus pneumoniae by children in day-care centers during an intervention program in Malmo, Sweden. Pediatr Infect Dis J. 2001;20(12):1144–9.
Rackham D, Ray S, Franks A, Bielak K, Pinn T. Community-Associated methicillin-resistant Staphylococcus aureus nasal carriage in a college student athlete population. Clin J Sport Med. 2010;20(3):185–8.
Groat EF, Williams NJ, Pinchbeck G, Simpson A, Schmidt VM. Canine raw meat diets and antimicrobial resistant E. coli: is there a link? BSAVA Congr Proc 2016 [Internet]. 2018;57:540–540. https://doi.org/10.22233/9781910443446.69.5.
Olkkola S, Kovanen S, Roine J, Hanninen M-L, Hielm-Bjorkman A, Kivisto R. Population genetics and antimicrobial susceptibility of canine campylobacter isolates collected before and after a raw feeding experiment. PLoS ONE. 2015;10(7):e0132660.
Fukuda A, Usui M, Okamura M, Dong-Liang H, Tamura Y. Role of flies in the maintenance of antimicrobial resistance in farm environments. Microb Drug Resist. 2019;25(1):127–32.
Barbarin AM, Hu B, Nachamkin I, Levy MZ. Colonization of Cimex lectularius with methicillin-resistant Staphylococcus aureus. Vol. 16, Environmental microbiology. England; 2014. p. 1222–4.
Lis DO, Pacha JZ, Idzik D. Methicillin resistance of airborne coagulase-negative staphylococci in homes of persons having contact with a hospital environment. Am J Infect Control. 2009;37(3):177–82.
Kennedy M, Ramsheh MY, Williams CML, Auty J, Haldar K, Abdulwhhab M, et al. Face mask sampling reveals antimicrobial resistance genes in exhaled aerosols from patients with chronic obstructive pulmonary disease and healthy volunteers. BMJ Open Respir Res. 2018;5(1):e000321.
Thom KA, Johnson JK, Lee MS, Harris AD. Environmental contamination because of multidrug-resistant Acinetobacter baumannii surrounding colonized or infected patients. Am J Infect Control. 2011;39(9):711–5.
Bures S, Fishbain JT, Uyehara CFT, Parker JM, Berg BW. Computer keyboards and faucet handles as reservoirs of nosocomial pathogens in the intensive care unit. Am J Infect Control. 2000;28(6):465–71.
Shahbazian JH, Hahn PD, Ludwig S, Ferguson J, Baron P, Christ A, et al. Multidrug and mupirocin resistance in environmental methicillin-resistant Staphylococcus aureus (MRSA) isolates from homes of people diagnosed with community-onset MRSA infection. Appl Environ Microbiol. 2017;83(22).
Levin AS, Gobara S, Mendes CMF, Cursino MR, Sinto S. Environmental contamination by multidrug-resistant Acinetobacter baumannii in an intensive care unit. Infect Control Hosp Epidemiol. 2001;22(11):717–20.
Zimbudzi E, Stuart RL, Korman TM, Kotsanas D. Contamination of renal patients’ hospital chart covers with vancomycin– resistant enterococci: handle with care. Australas Med J. 2011;4(10):538–41.
Chalfine A, Kitzis M-D, Bezie Y, Benali A, Perniceni L, Nguyen J-C, et al. Ten-year decrease of acquired methicillin-resistant Staphylococcus aureus (MRSA) bacteremia at a single institution: the result of a multifaceted program combining cross-transmission prevention and antimicrobial stewardship. Antimicrob Resist Infect Control. 2012;1(1):18.
Duggal S, Kaur N, Hans C. An investigation of MRSA from the burns ward: the importance of hand hygiene. J Clin Diagnostic Res [Internet]. 2011;5(3):476–9.
Batra R, Cooper BS, Whiteley C, Patel AK, Wyncoll D, Edgeworth JD. Efficacy and limitation of a chlorhexidine-based decolonization strategy in preventing transmission of methicillin-resistant Staphylococcus aureus in an intensive care unit. Clin Infect Dis [Internet]. 2010;50(2):210–7. https://doi.org/10.1086/648717.
D’Agata EM, Horn MA, Webb GF. The impact of persistent gastrointestinal colonization on the transmission dynamics of vancomycin-resistant enterococci. J Infect Dis. 2002;185(6):766–73.
Perencevich EN, Fisman DN, Lipsitch M, Harris AD, Morris JG Jr, Smith DL. Projected benefits of active surveillance for vancomycin-resistant enterococci in intensive care units. Clin Infect Dis. 2004;38(8):1108–15.
Montecalvo MA, Jarvis WR, Uman J, Shay DK, Petrullo C, Rodney K, et al. Infection-control measures reduce transmission of vancomycin-resistant enterococci in an endemic setting. Ann Intern Med. 1999;131(4):269–72.
Coleman BL, Louie M, Salvadori MI, McEwen SA, Neumann N, Sibley K, et al. Contamination of Canadian private drinking water sources with antimicrobial resistant Escherichia coli. Water Res. 2013;47(9):3026–36.
Caudell MA, Mair C, Subbiah M, Matthews L, Quinlan RJ, Quinlan MB, et al. Identification of risk factors associated with carriage of resistant Escherichia coli in three culturally diverse ethnic groups in Tanzania: a biological and socioeconomic analysis. Lancet Planet Heal. 2018;2(11):e489–97.
Moodley A, Latronico F, Guardabassi L. Experimental colonization of pigs with methicillin-resistant Staphylococcus aureus (MRSA): insights into the colonization and transmission of livestock-associated MRSA. Epidemiol Infect. 2011;139(10):1594–600.
Corrò M, Skarin J, Börjesson S, Rota A. Occurrence and characterization of methicillin-resistant Staphylococcus pseudintermedius in successive parturitions of bitches and their puppies in two kennels in Italy. BMC Vet Res. 2018;14(1):308.
Tameliene R, Barčaite E, Stoniene D, Buinauskiene J, Markuniene E, Kudrevičiene A, et al. Escherichia coli colonization in neonates: prevalence, perinatal transmission, antimicrobial susceptibility, and risk factors. Med. 2012;48(2):71–6.
Denkel LA, Schwab F, Kola A, Leistner R, Garten L, von Weizsäcker K, et al. The mother as most important risk factor for colonization of very low birth weight (VLBW) infants with extended-spectrum b-lactamase-producing Enterobacteriaceae (ESBL-E). J Antimicrob Chemother [Internet]. 2014;69(8):2230–7. https://doi.org/10.1093/jac/dku097.
Kunze M, Ziegler A, Fluegge K, Hentschel R, Proempeler H, Berner R. Colonization, serotypes and transmission rates of group B streptococci in pregnant women and their infants born at a single University Center in Germany. J Perinat Med [Internet]. 2011;39(4):417–22. https://doi.org/10.1515/jpm.2011.037.
Bourgeois-Nicolaos N, Lucet JC, Daubié C, Benchaba F, Rajguru M, Ruimy R, et al. Maternal vaginal colonisation by Staphylococcus aureus and newborn acquisition at delivery. Paediatr Perinat Epidemiol [Internet]. 2010;24(5):488–91. https://doi.org/10.1111/j.1365-3016.2010.01139.x.
Chua KB, Ngeow YF, Ng KB, Chye JK, Lim CT. Ureaplasma urealyticum and Mycoplasma hominis isolation from cervical secretions of pregnant women and nasopharyngeal secretions of their babies at delivery. Singapore Med J [Internet]. 1998;39(7):300–2.
Eren A, Küçükercan M, Oǧuzoǧlu N, Ünal N, Karateke A. The carriage of group B streptococci in Turkish pregnant women and its transmission rate in newborns and serotype distribution. Turk J Pediatr. 2005;47(1):28–33.
Facchinetti F, Piccinini F, Mordini B, Volpe A. Chlorhexidine vaginal flushings versus systemic ampicillin in the prevention of vertical transmission of neonatal group B streptococcus, at term. J Matern Neonatal Med [Internet]. 2002;11(2):84–8.
Jiménez-Rámila C, López-Cerero L, Aguilar Martín MV, Vera Martín C, Serrano L, Pascual A, et al. Vagino-rectal colonization and maternal–neonatal transmission of Enterobacteriaceae producing extended-spectrum β-lactamases or carbapenemases: a cross-sectional study. J Hosp Infect. 2019;101(2):167–74.
Kadanali A, Altoparlak U, Kadanali S. Maternal carriage and neonatal colonisation of group B streptococcus in eastern Turkey: prevalence, risk factors and antimicrobial resistance. Int J Clin Pr. 2005;59(4):437–40.
Kunze M, Zumstein K, Markfeld-Erol F, Elling R, Lander F, Prömpeler H, et al. Comparison of pre- and intrapartum screening of group B streptococci and adherence to screening guidelines: a cohort study. Eur J Pediatr [Internet]. 2015;174(6):827–35. https://doi.org/10.1007/s00431-015-2548-y.
Strus M, Pawlik D, Brzychczy-Włoch M, Gosiewski T, Rytlewski K, Lauterbach R, et al. Group B streptococcus colonization of pregnant women and their children observed on obstetric and neonatal wards of the University hospital in Krakow, Poland. J Med Microbiol. 2009;58(2):228–33.
Kothari C, Gaind R, Singh LC, Sinha A, Kumari V, Arya S, et al. Community acquisition of beta-lactamase producing Enterobacteriaceae in neonatal gut. BMC Microbiol. 2013;17(13):136.
Morel AS, Wu F, Della-Latta P, Cronquist A, Rubenstein D, Saiman L. Nosocomial transmission of methicillin-resistant Staphylococcus aureus from a mother to her preterm quadruplet infants. Am J Infect Control. 2002;30(3):170–3.
Peretz A, Skuratovsky A, Khabra E, Adler A, Pastukh N, Barak S, et al. Peripartum maternal transmission of extended-spectrum β-lactamase organism to newborn infants. Diagn Microbiol Infect Dis [Internet]. 2017;87(2):168–71. https://doi.org/10.1016/j.diagmicrobio.2016.11.004.
Kurita H, Kurashina K, Honda T. Nosocomial transmission of methicillin-resistant Staphylococcus aureus via the surfaces of the dental operatory. Br Dent J. 2006;201(5):297–300.
Zhou ZY, Hu BJ, Gao XD, Bao R, Chen M, Li HY. Sources of sporadic Pseudomonas aeruginosa colonizations/infections in surgical ICUs: association with contaminated sink trap. J Infect Chemother [Internet]. 2016;22(7):450–5. https://doi.org/10.1016/j.jiac.2016.03.016.
Rubini S, Ravaioli C, Previato S, D’Incau M, Tassinari M, Guidi E, et al. Prevalence of Salmonella strains in wild animals from a highly populated area of north-eastern Italy. Ann Ist Super Sanita. 2016;52(2):277–80.
Wielders CCH, van Hoek AHAM, Hengeveld PD, Veenman C, Dierikx CM, Zomer TP, et al. Extended-spectrum β-lactamase- and pAmpC-producing Enterobacteriaceae among the general population in a livestock-dense area. Clin Microbiol Infect [Internet]. 2017;23(2):120.e1-120.e8. https://doi.org/10.1016/j.cmi.2016.10.013.
Reynaga E, Torres C, Garcia-Nuñez M, Navarro M, Vilamala A, Puigoriol E, et al. Clinical impact and prevalence of MRSA CC398 and differences between MRSA-TetR and MRSA-TetS in an area of Spain with a high density of pig farming: a prospective cohort study. Clin Microbiol Infect. 2017;23(9):678.e1-678.e4.
Diercke M, Rocker D, Köck R, Pulz M. Livestock-associated methicillin-resistant Staphylococcus aureus in patients admitted to hospitals in the Ems-Dollart-Region, 2011. Int J Med Microbiol [Internet]. 2012;302:138–9. https://doi.org/10.1016/j.ijmm.2012.08.002.
Founou LL, Founou RC, Essack SY, Djoko CF. Mannitol-fermenting methicillin-resistant staphylococci (MRS) in pig abattoirs in Cameroon and South Africa: a serious food safety threat. Int J Food Microbiol. 2018;285:50–60.
Ho J, O’Donoghue MM, Boost MV. Occupational exposure to raw meat: a newly-recognized risk factor for Staphylococcus aureus nasal colonization amongst food handlers. Int J Hyg Environ Health. 2014;217(2–3):347–53.
Jayaweera JAAS, Kumbukgolla WW. Antibiotic resistance patterns of methicillin-resistant Staphylococcus aureus (MRSA) isolated from livestock and associated farmers in Anuradhapura, Sri Lanka. Germs. 2017;7(3):132–9.
Moodley A, Nightingale EC, Stegger M, Nielsen SS, Skov RL, Guardabassi L. High risk for nasal carriage of methicillin-resistant Staphylococcus aureus among Danish veterinary practitioners. Scand J Work Environ Heal. 2008;34(2):151–7.
Rasamiravaka T, Nirinarimanana AJ, Rasamindrakotroka A. Evaluation of methicillin-resistant Staphylococcus aureus nasal carriage in malagasy veterinary students: MRSA nasal carriage inmalagasy veterinary students. Afr J Clin Exp Microbiol [Internet]. 2016;17(4):250–5. https://doi.org/10.4314/ajcem.v17i4.5.
Rosenberg Goldstein RE, Micallef SA, Gibbs SG, He X, George A, Sapkota A, et al. Occupational exposure to Staphylococcus aureus and Enterococcus spp. among spray irrigation workers using reclaimed water. Int J Environ Res Public Health. 2014;11(4):4340–55.
van Rijen MM, Van Keulen PH, Kluytmans JA. Increase in a Dutch hospital of methicillin-resistant Staphylococcus aureus related to animal farming. Clin Infect Dis. 2008;46(2):261–3.
Ye X, Fan Y, Wang X, Liu W, Yu H, Zhou J, et al. Livestock-associated methicillin and multidrug resistant S. aureus in humans is associated with occupational pig contact, not pet contact. Sci Rep. 2016;6(6):19184.
Ye X, Wang X, Fan Y, Peng Y, Li L, Li S, et al. Genotypic and phenotypic markers of livestock-associated methicillin-resistant Staphylococcus aureus CC9 in humans. Appl Env Microbiol. 2016;82(13):3892–9.
Price LB, Graham JP, Lackey LG, Roess A, Vailes R, Silbergeld E. Elevated risk of carrying gentamicin-resistant Escherichia coli among U.S. poultry workers. Environ Health Perspect. 2007;115(12):1738–42.
Trung NV, Matamoros S, Carrique-Mas JJ, Nghia NH, Nhung NT, Chieu TTB, et al. Zoonotic transmission of mcr-1 colistin resistance gene from small-scale poultry farms. Vietnam Emerg Infect Dis. 2017;23(3):529–32.
Moser KA, Zhang L, Spicknall I, Braykov NP, Levy K, Marrs CF, et al. The role of mobile genetic elements in the spread of antimicrobial-resistant Escherichia coli from chickens to humans in small-scale production poultry operations in rural ecuador. Am J Epidemiol. 2018;187(3):558–67.
Wardyn SE, Forshey BM, Farina SA, Kates AE, Nair R, Quick MK, et al. Swine farming is a risk factor for infection with and high prevalence of carriage of multidrug-resistant Staphylococcus aureus. Clin Infect Dis. 2015;61(1):59–66.
Rinsky JL, Nadimpalli M, Wing S, Hall D, Baron D, Price LB, et al. Livestock-associated methicillin and multidrug resistant Staphylococcus aureus is present among industrial, not antibiotic-free livestock operation workers in North Carolina. PLoS ONE. 2013;8(7):e67641.
Hatcher SM, Rhodes SM, Stewart JR, Silbergeld E, Pisanic N, Larsen J, et al. The prevalence of antibiotic-resistant Staphylococcus aureus nasal carriage among industrial hog operation workers, community residents, and children living in their households: North Carolina, USA. Env Heal Perspect. 2017;125(4):560–9.
Borgen K, Simonsen GS, Sundsfjord A, Wasteson Y, Olsvik O, Kruse H. Continuing high prevalence of VanA-type vancomycin-resistant enterococci on Norwegian poultry farms three years after avoparcin was banned. J Appl Microbiol. 2000;89(3):478–85.
Fischer J, Hille K, Mellmann A, Schaumburg F, Kreienbrock L, Koeck R. Nasal colonization of pig-exposed persons with Enterobacteriaceae and associated antimicrobial resistance. Int J Med Microbiol [Internet]. 2015;305:93. https://doi.org/10.3390/pathogens8010010.
Frana TS, Beahm AR, Hanson BM, Kinyon JM, Layman LL, Karriker LA, et al. Isolation and characterization of methicillin-resistant Staphylococcus aureus from pork farms and visiting veterinary students. PLoS ONE. 2013;8(1): e53738. https://doi.org/10.1371/journal.pone.0053738.
Wulf MWH, Sørum M, Van Nes A, Skov R, Melchers WJG, Klaassen CHW, et al. Prevalence of methicillin-resistant Staphylococcus aureus among veterinarians: an international study. Clin Microbiol Infect [Internet]. 2008;14(1):29–34. https://doi.org/10.1111/j.1469-0691.2007.01873.x.
Fang HW, Chiang PH, Huang YC. Livestock-associated methicillin-resistant Staphylococcus aureus ST9 in pigs and related personnel in Taiwan. PLoS One [Internet]. 2014. https://doi.org/10.1371/journal.pone.0088826.
Khanna T, Friendship R, Dewey C, Weese JS. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet Microbiol. 2008;128(3–4):298–303.
Witte W, Layer F. Emergence of caMRSA infections in humans caused by MRSA CC398 which has its main reservoir in animals. Int J Med Microbiol [Internet]. 2009;299:105. https://doi.org/10.2807/1560-7917.ES.2015.20.37.30021.
Drougka E, Foka A, Koutinas CK, Jelastopulu E, Giormezis N, Farmaki O, et al. Interspecies spread of Staphylococcus aureus clones among companion animals and human close contacts in a veterinary teaching hospital. A cross-sectional study in Greece. Prev Vet Med. 2016;126:190–8.
Oppliger A, Moreillon P, Charrière N, Giddey M, Morisset D, Sakwinska O. Antimicrobial resistance of Staphylococcus aureus strains acquired by pig farmers from pigs. Appl Environ Microbiol. 2012;78(22):8010–4.
Nakane K, Kawamura K, Goto K, Arakawa Y. Long-term colonization by blaCTX-M-harboring Escherichia coli in healthy Japanese people engaged in food handling. Appl Environ Microbiol. 2016;82(6):1818–27.
Castro A, Santos C, Meireles H, Silva J, Teixeira P. Food handlers as potential sources of dissemination of virulent strains of Staphylococcus aureus in the community. J Infect Public Health. 2016;9(2):153–60.
Zurfluh K, Stephan R, Widmer A, Poirel L, Nordmann P, Nüesch HJ, et al. Screening for fecal carriage of MCR-producing Enterobacteriaceae in healthy humans and primary care patients. Antimicrob Resist Infect Control [Internet]. 2017. https://doi.org/10.1186/s13756-017-0186-z.
Cuny C, Arnold P, Hermes J, Eckmanns T, Mehraj J, Schoenfelder S, et al. Occurrence of cfr-mediated multiresistance in staphylococci from veal calves and pigs, from humans at the corresponding farms, and from veterinarians and their family members. Vet Microbiol. 2017;200:88–94.
Brennan GI, Abbott Y, Burns A, Leonard F, McManus BA, O’Connell B, et al. The emergence and spread of multiple livestock-associated clonal complex 398 methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains among animals and humans in the Republic of Ireland, 2010–2014. PLoS ONE. 2016;11(2):e0149396.
Lozano C, Aspiroz C, Lasarte JJ, Gomez-Sanz E, Zarazaga M, Torres C. Dynamic of nasal colonization by methicillin-resistant Staphylococcus aureus ST398 and ST1 after mupirocin treatment in a family in close contact with pigs. Comp Immunol Microbiol Infect Dis. 2011;34(1):e1-7.
Tzavaras I, Siarkou VI, Zdragas A, Kotzamanidis C, Vafeas G, Bourtzi-Hatzopoulou E, et al. Diversity of vanA-type vancomycin-resistant Enterococcus faecium isolated from broilers, poultry slaughterers and hospitalized humans in Greece. J Antimicrob Chemother. 2012;67(8):1811–8.
Kock R, Schaumburg F, Mellmann A, Koksal M, Jurke A, Becker K, et al. Livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) as causes of human infection and colonization in Germany. PLoS ONE. 2013;8(2):e55040.
Wang J, Lin D-C, Guo X-M, Wei H-K, Liu X-Q, Chen X-J, et al. Distribution of the multidrug resistance gene cfr in Staphylococcus isolates from pigs, workers, and the environment of a hog market and a Slaughterhouse in Guangzhou. China Foodborne Pathog Dis. 2015;12(7):598–605.
Mularoni A, Bertani A, Vizzini G, Gona F, Campanella M, Spada M, et al. Outcome of transplantation using organs from donors infected or colonized with carbapenem-resistant gram-negative bacteria. Am J Transplant. 2015;15(10):2674–82.
Goldberg E, Bishara J, Lev S, Singer P, Cohen J. Organ transplantation from a donor colonized with a multidrug-resistant organism: a case report. Transpl Infect Dis. 2012;14(3):296–9.
Cui M, Wu C, Zhang J, Zhang C, Li R, Wai-Chi Chan E, et al. Distinct mechanisms of acquisition of mcr-1 -bearing plasmid by Salmonella strains recovered from animals and food samples. Sci Rep. 2017;7(1):13199.
Mork RL, Hogan PG, Muenks CE, Boyle MG, Thompson RM, Morelli JJ, et al. Comprehensive modeling reveals proximity, seasonality, and hygiene practices as key determinants of MRSA colonization in exposed households. Pediatr Res. 2018;84(5):668–76.
Gandolfi-Decristophoris P, De Benedetti A, Petignat C, Attinger M, Guillaume J, Fiebig L, et al. Evaluation of pet contact as a risk factor for carriage of multidrug-resistant staphylococci in nursing home residents. Am J Infect Control [Internet]. 2012;40(2):128–33. https://doi.org/10.1016/j.ajic.2011.04.007.
Morita JE, Fujioka RS, Tice AD, Berestecky J, Sato D, Seifried SE, et al. Survey of methicillin-resistant Staphylococcus aureus (MRSA) carriage in healthy college students. Hawai’i Hawaii Med J. 2007;66(8):213–5.
Loncaric I, Brunthaler R, Spergser J. Suspected goat-to-human transmission of methicillin-resistant Staphylococcus aureus sequence type 398. J Clin Microbiol [Internet]. 2013;51(5):1625–6. https://doi.org/10.1128/JCM.03052-12.
Morris DO, Lautenbach E, Zaoutis T, Leckerman K, Edelstein PH, Rankin SC. Potential for pet animals to harbour methicillin-resistant Staphylococcus aureus when residing with human MRSA patients. Zoonoses Public Heal. 2012;59(4):286–93.
Boost MV, O’Donoghue MM, James A. Prevalence of Staphylococcus aureus carriage among dogs and their owners. Epidemiol Infect. 2008;136(7):953–64.
Boehmer T, Vogler AJ, Thomas A, Sauer S, Hergenroether M, Straubinger RK, et al. Phenotypic characterization and whole genome analysis of extended-spectrum betalactamase-producing bacteria isolated from dogs in Germany. PLoS One [Internet]. 2018. https://doi.org/10.1371/journal.pone.0206252.
Damborg P, Top J, Hendrickx APA, Dawson S, Willems RJL, Guardabassi L. Dogs are a reservoir of ampicillin-resistant Enterococcus faecium lineages associated with human infections. Appl Environ Microbiol. 2009;75(8):2360–5.
Van den Eede A, Martens A, Floré K, Denis O, Gasthuys F, Haesebrouck F, et al. MRSA carriage in the equine community: an investigation of horse-caretaker couples. Vet Microbiol. 2013;163(3–4):313–8.
Walther B, Hermes J, Cuny C, Wieler LH, Vincze S, Elnaga YA, et al. Sharing more than friendship—nasal colonization with coagulase-positive staphylococci (CPS) and co-habitation aspects of dogs and their owners. PLoS One [Internet]. 2012. https://doi.org/10.1371/journal.pone.0035197.
Shorman M, Al-Tawfiq JA. Risk factors associated with vancomycin-resistant enterococcus in intensive care unit settings in Saudi Arabia. Interdiscip Perspect Infect Dis. 2013;2013:369674.
Nseir S, Blazejewski C, Lubret R, Wallet F, Courcol R, Durocher A. Risk of acquiring multidrug-resistant Gram-negative bacilli from prior room occupants in the intensive care unit. Clin Microbiol Infect [Internet]. 2011;17(8):1201–8. https://doi.org/10.1111/j.1469-0691.2010.03238.x.
Morales Barroso I, López-Cerero L, Navarro MD, Gutiérrez-Gutiérrez B, Pascual A, Rodríguez-Baño J. Intestinal colonization due to Escherichia coli ST131: risk factors and prevalence. Antimicrob Resist Infect Control [Internet]. 2018. https://doi.org/10.1186/s13756-018-0427-9.
Torres-Gonzalez P, Cervera-Hernandez ME, Niembro-Ortega MD, Leal-Vega F, Cruz-Hervert LP, Garcia-Garcia L, et al. Factors associated to prevalence and incidence of Carbapenem-resistant Enterobacteriaceae fecal carriage: a cohort study in a Mexican Tertiary Care Hospital. PLoS ONE. 2015;10(10):e0139883.
Ben-David D, Masarwa S, Navon-Venezia S, Mishali H, Fridental I, Rubinovitch B, et al. Carbapenem-resistant Klebsiella pneumoniae in post-acute-care facilities in Israel. Infect Control Hosp Epidemiol. 2011;32(9):845–53.
Papadimitriou-Olivgeris M, Drougka E, Fligou F, Kolonitsiou F, Liakopoulos A, Dodou V, et al. Risk factors for enterococcal infection and colonization by vancomycin-resistant enterococci in critically ill patients. Infection. 2014;42(6):1013–22.
Jiménez-Truque N, Saye EJ, Soper N, Saville BR, Thomsen I, Edwards KM, et al. Association between contact sports and colonization with Staphylococcus aureus in a prospective cohort of collegiate athletes. Sport Med [Internet]. 2017;47(5):1011–9. https://doi.org/10.1007/s40279-016-0618-6.
Fossi Djembi L, Hodille E, Chomat-Jaboulay S, Coudrais S, De Santis N, Gardes S, et al. Factors associated with Vancomycin-resistant Enterococcus acquisition during a large outbreak. J Infect Public Health. 2017;10(2):185–90.
Ulstad CR, Solheim M, Berg S, Lindbæk M, Dahle UR, Wester AL. Carriage of ESBL/AmpC-producing or ciprofloxacin non-susceptible Escherichia coli and Klebsiella spp. in healthy people in Norway. Antimicrob Resist Infect Control. 2016;5(1):57.
Vading M, Kabir MH, Kalin M, Iversen A, Wiklund S, Nauclér P, et al. Frequent acquisition of low-virulence strains of ESBL-producing Escherichia coli in travellers. J Antimicrob Chemother. 2016;71(12):3548–55.
Peirano G, Gregson DB, Kuhn S, Erkooi OG, Nobrega DB, Pitout JDD. Rates of colonization with extended-spectrum beta-lactamase-producing Escherichia coli in Canadian travellers returning from South Asia: a cross-sectional assessment. C Open. 2017;5(4):E850-e855.
Macfadden DR, Bogoch II, Brownstein JS, Daneman N, Fisman D, German M, et al. A passage from India: association between air traffic and reported cases of New Delhi Metallo-beta-lactamase 1 from 2007 to 2012. Travel Med Infect Dis. 2015;13(4):295–9.
Epelboin L, Robert J, Tsyrina-Kouyoumdjian E, Laouira S, Meyssonnier V, Caumes E. High rate of multidrug-resistant gram-negative bacilli carriage and infection in hospitalized returning travelers: a cross-sectional cohort study. J Travel Med. 2015;22(5):292–9.
Lorme F, Maataoui N, Rondinaud E, Esposito-Farèse M, Clermont O, Ruppe E, et al. Acquisition of plasmid-mediated cephalosporinase producing Enterobacteriaceae after a travel to the tropics. PLoS ONE. 2018;13(12):e0206909.
Nurjadi D, Friedrich-Jänicke B, Schäfer J, Van Genderen PJJ, Goorhuis A, Perignon A, et al. Skin and soft tissue infections in intercontinental travellers and the import of multi-resistant Staphylococcus aureus to Europe. Clin Microbiol Infect. 2015;21(6):567.e1-567.e10.
Ostholm-Balkhed A, Tarnberg M, Nilsson M, Nilsson LE, Hanberger H, Hallgren A, et al. Travel-associated faecal colonization with ESBL-producing Enterobacteriaceae: incidence and risk factors. J Antimicrob Chemother. 2013;68(9):2144–53.
Yaita K, Aoki K, Suzuki T, Nakaharai K, Yoshimura Y, Harada S, et al. Epidemiology of extended-spectrum β-lactamase producing Escherichia coli in the stools of returning Japanese travelers, and the risk factors for colonization. PLoS One [Internet]. 2014. https://doi.org/10.1371/journal.pone.0098000.
Sannes MR, Belongia EA, Kieke B, Smith K, Kieke A, Vandermause M, et al. Predictors of antimicrobial-resistant Escherichia coli in the feces of vegetarians and newly hospitalized adults in minnesota and wisconsin. J Infect Dis. 2008;197(3):430–4.
Islam S, Selvarangan R, Kanwar N, McHenry R, Chappell JD, Halasa N, et al. Intestinal carriage of third-generation cephalosporin-resistant and extended-spectrum β-lactamase-producing enterobacteriaceae in healthy US children. J Pediatric Infect Dis Soc. 2018;7(3):234–40.
Coleman BL, Salvadori MI, McGeer AJ, Sibley KA, Neumann NF, Bondy SJ, et al. The role of drinking water in the transmission of antimicrobial-resistant E. coli. Epidemiol Infect [Internet]. 2012;140(4):633–42. https://doi.org/10.1017/S0950268811001038.
Tängdén T, Cars O, Melhus Å, Löwdin E. Foreign travel is a major risk factor for colonization with Escherichia coli producing CTX-M-type extended-spectrum β-lactamases: a prospective study with Swedish volunteers. Antimicrob Agents Chemother [Internet]. 2010;54(9):3564–8. https://doi.org/10.1128/AAC.00220-10.
Pires J, Kuenzli E, Kasraian S, Tinguely R, Furrer H, Hilty M, et al. Polyclonal intestinal colonization with extended-spectrum cephalosporin-resistant enterobacteriaceae upon traveling to India. Front Microbiol. 2016;7(JUL):1069.
Lausch KR, Fuursted K, Larsen CS, Storgaard M. Colonisation with multi-resistant Enterobacteriaceae in hospitalised Danish patients with a history of recent travel: a cross-sectional study. Travel Med Infect Dis. 2013;11(5):320–3.
Arcilla MS, van Hattem JM, Haverkate MR, Bootsma MCJ, van Genderen PJJ, Goorhuis A, et al. Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect Dis. 2017;17(1):78–85.
Blyth DM, Mende K, Maranich AM, Beckius ML, Harnisch KA, Rosemann CA, et al. Antimicrobial resistance acquisition after international travel in U.S. travelers. Trop Dis Travel Med Vaccines. 2016;2:4.
Bevan ER, McNally A, Thomas CM, Piddock LJV, Hawkey PM. Acquisition and loss of CTX-M-producing and non-producing Escherichia coli in the fecal microbiome of travelers to South Asia. MBio. 2018;9(6).
Lubbert C, Straube L, Stein C, Makarewicz O, Schubert S, Mossner J, et al. Colonization with extended-spectrum beta-lactamase-producing and carbapenemase-producing Enterobacteriaceae in international travelers returning to Germany. Int J Med Microbiol. 2015;305(1):148–56.
Nakayama T, Kumeda Y, Kawahara R, Yamaguchi T, Yamamoto Y. Carriage of colistin-resistant, extended-spectrum β-lactamase-producing Escherichia coli harboring the mcr-1 resistance gene after short-term international travel to Vietnam. Infect Drug Resist [Internet]. 2018;11:391–5. https://doi.org/10.2147/IDR.S153178.
Valverde A, Turrientes MC, Norman F, Pérez Molina JA, Coque T, Baquero F, et al. Intestinal Colonisation with multidrug-resistant Enterobacteriaceae in travellers, immigrants and “visiting friends and relatives”: dominance of E. coli producing CTX-M enzymes. Clin Microbiol Infect [Internet]. 2012;18:763–4. https://doi.org/10.1111/j.1469-0691.2012.03802.x.
Johnning A, Kristiansson E, Angelin M, Marathe N, Shouche YS, Johansson A, et al. Quinolone resistance mutations in the faecal microbiota of Swedish travellers to India. BMC Microbiol. 2015;15(1):15–85.
Sun Q, Tarnberg M, Zhao L, Stalsby Lundborg C, Song Y, Grape M, et al. Varying high levels of faecal carriage of extended-spectrum beta-lactamase producing Enterobacteriaceae in rural villages in Shandong, China: implications for global health. PLoS ONE. 2014;9(11):e113121.
O’Flaherty E, Solimini AG, Pantanella F, De Giusti M, Cummins E. Human exposure to antibiotic resistant-Escherichia coli through irrigated lettuce. Environ Int. 2019;122:270–80.
Dekić S, Klobučar G, Ivanković T, Zanella D, Vucić M, Bourdineaud JP, et al. Emerging human pathogen Acinetobacter baumannii in the natural aquatic environment: a public health risk? Int J Environ Health Res. 2018;28(3):315–22.
Goldstein RR, Kleinfelter L, He X, Micallef SA, George A, Gibbs SG, et al. Higher prevalence of coagulase-negative staphylococci carriage among reclaimed water spray irrigators. Sci Total Environ. 2017;595:35–40.
Leonard AFC, Zhang L, Balfour AJ, Garside R, Hawkey PM, Murray AK, et al. Exposure to and colonisation by antibiotic-resistant E. coli in UK coastal water users: environmental surveillance, exposure assessment, and epidemiological study (Beach Bum Survey). Environ Int. 2018;114:326–33.
Yang F, Huang L, Li L, Yang Y, Mao D, Luo Y. Discharge of KPC-2 genes from the WWTPs contributed to their enriched abundance in the receiving river. Sci Total Environ [Internet]. 2017;581:136–43. https://doi.org/10.1016/j.scitotenv.2016.12.063.
Acknowledgements
None.
Funding
This research was part of the Risk and Disease burden of Antimicrobial Resistance (RaDAR) project, which was funded through the One Health European Joint Programme by the EU’s Horizon-2020 Research and Innovation Programme (Grant 773830) (https://ec.europa.eu/info/research-and-innovation_en).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. M.C.J.B. and N.G.G. were funded through this Grant.
Author information
Authors and Affiliations
Contributions
NGG, MCJB and MJMB conceptualized and designed the study. NGG undertook the literature review, extracted the data, analysed and drafted the manuscript. MCJB assisted in the literature review and drafting of the manuscript. MJMB and MCJB reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participants
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Supporting information including the search terms, Prisma checklist, frequency tables and a list of which studies were included in the review.
Additional file 2.
Transmission data identified in the systematic review by Godijk et al.
Additional file 3.
R Script of the systematic review by Godijk et al.
Additional file 4.
R script of the forest plots for the systematic review of Godijk et al.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Godijk, N.G., Bootsma, M.C.J. & Bonten, M.J.M. Transmission routes of antibiotic resistant bacteria: a systematic review. BMC Infect Dis 22, 482 (2022). https://doi.org/10.1186/s12879-022-07360-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-022-07360-z