Abstract
Background
Joint models for longitudinal and time-to-event data are commonly used to simultaneously analyse correlated data in single study cases. Synthesis of evidence from multiple studies using meta-analysis is a natural next step but its feasibility depends heavily on the standard of reporting of joint models in the medical literature. During this review we aim to assess the current standard of reporting of joint models applied in the literature, and to determine whether current reporting standards would allow or hinder future aggregate data meta-analyses of model results.
Methods
We undertook a literature review of non-methodological studies that involved joint modelling of longitudinal and time-to-event medical data. Study characteristics were extracted and an assessment of whether separate meta-analyses for longitudinal, time-to-event and association parameters were possible was made.
Results
The 65 studies identified used a wide range of joint modelling methods in a selection of software. Identified studies concerned a variety of disease areas. The majority of studies reported adequate information to conduct a meta-analysis (67.7% for longitudinal parameter aggregate data meta-analysis, 69.2% for time-to-event parameter aggregate data meta-analysis, 76.9% for association parameter aggregate data meta-analysis). In some cases model structure was difficult to ascertain from the published reports.
Conclusions
Whilst extraction of sufficient information to permit meta-analyses was possible in a majority of cases, the standard of reporting of joint models should be maintained and improved. Recommendations for future practice include clear statement of model structure, of values of estimated parameters, of software used and of statistical methods applied.
Similar content being viewed by others
Background
Joint modelling of longitudinal and time-to-event data is an area of increasing research [1–3], which allows the simultaneous modelling of a longitudinal (repeatedly measured over time) outcome such as weekly biomarker measurements, and a time-to-event (survival) outcome such as time to death. The model consists of two sub-models; a longitudinal sub-model (such as a linear mixed effects model) and a time-to-event sub-model (such as a cox proportional hazards models) which are linked using an association structure that quantifies the relationship between the outcomes of interest.
Within a single study, joint models have the potential to reduce parameter estimate bias, account for dropout in longitudinal studies and enable the inclusion of longitudinal covariates measured with error in time-to-event models [1, 4]. These qualities often make joint models preferable to separate longitudinal or time-to-event analyses. Joint models have been applied in the literature to investigate links between biomarkers and certain disease events (e.g. in cancer studies), and to account for informative study dropout.
Glass 1976 [5] defined meta-analysis (MA) as the statistical analysis or pooling of results from separate studies. Such analyses can increase power and precision compared to original studies, or answer questions additional to those originally posed [6]. MA can be performed on the original Individual Participant Data (IPD), or on the study level results (published in the literature or obtained from authors) termed Aggregate Data (AD). Overviews of MA methodology can be found in Whitehead 2002 [7] and in the Cochrane handbook [6].
Whilst the benefits of joint modelling methods for individual studies are well established, little attention has been given towards the potential value of pooling estimates across similar studies in an aggregate data meta-analysis (AD-MA) of joint models. However before an AD-MA using published data can be undertaken, relevant studies must be identified, and the necessary information must be extracted. We aim to investigate the reporting of joint longitudinal and time-to-event models applied to real medical data in the literature to establish whether current reporting practices would allow sufficient data to be extracted to undertake AD-MA.
Methods
Identification of papers
We performed our systematic review in accordance to the guidelines of the Preferred Reporting of Items for Systematic Reviews and Meta-Analyses (PRISMA) [8]. We searched the Medline, Pubmed and Scopus datasets for studies using joint models for longitudinal and time-to-event data to analyse medical data (search strategies available in the Additional file 1).
Papers mentioning joint models for longitudinal (or repeated measures over time) data and time-to-event (or survival, event time or event history) data were identified. Duplicates were identified and removed. Abstracts and keywords were then examined, and irrelevant papers were removed. Examples of disregarded papers include papers modelling body joints, papers discussing joint models as a future extension or alternative to methods used, or papers using two stage approaches rather than simultaneous estimation of the longitudinal and time-to-event sub-models. Papers not relating to medical or biostatistical datasets were discarded (e.g. data from plant or animal subjects except from modelling of human diseases input into animal hosts). Additionally papers involving repeated measures over space rather than time were discarded (e.g. repeated measures across tumour sites). If study relevancy was unclear from the abstract, the full text was obtained and viewed after which the study was included or discarded.
Any retained papers were sorted into an applied and a methodological group. Some methodology papers presented results from application to example datasets. These were considered reanalyses of data or demonstrations of methods rather than primary analyses to influence future practice. Also, methodological papers might be expected to better report results as their authors are experts in the area. The aim of this review was to assess how well joint models are reported in the general medical literature, so we focussed on the applied group only.
Data extraction
A blank data extraction form is presented in Additional file 2. During the investigation we refer to references identified as applying joint models to relevant datasets as studies. Other publications (e.g. those cited by studies) are referred to as papers. Information recorded from identified studies included publication year, author, journal, joint model type, sharing structure between the longitudinal and time-to-event sub-models, types of sub-models, Bayesian or frequentist methods, and software used. Disease area was recorded (with respect to the type of longitudinal and time-to-event data, for example studies modelling biomarkers in heart disease patients after a transplant operation were classed as transplant data).
The sources of the methods used were recorded. Specifically if the study developed methods specific to their dataset, “own methods developed” was recorded. If the study referenced specific papers as the source of the methods they used the papers referenced were recorded.
Availability of information required for a MA was also recorded (including the number of participants, significance level, and presence longitudinal, time-to-event and association parameters along with their precision estimates). The significance level used was identified through direct statement in the text, or specified confidence interval sizes on tables, graphs, or footnotes relating to the joint models fitted. For a MA to be considered possible, the number of participants and model coefficients had to be reported, with either a standard error, or a confidence interval with accompanying significance level.
We assume for an AD-MA of joint longitudinal and time-to-event models that a separate MA would be conducted for each of the longitudinal, the time-to-event and the association parameters from the identified studies. Consequently when identified studies were assessed for sufficient information to conduct a MA, they were assessed separately for longitudinal, for time-to-event and for association information. Ideally all three separate groups of MA would be conducted, however if insufficient information was reported only a subset of these MA might be feasible.
The reason for joint models use (see Henderson et al [9) may influence what information is presented in the study report. If joint models were used to account for informative dropout, the study might not report the time-to-event parameter information (although if the time-to-event endpoint is clinically defined then time-to-event estimates should be reported). Similarly if interest was to include a longitudinal variable measured with error as a time-variable covariate in a time-to-event model, the study may not clearly report longitudinal sub-model parameters. To investigate whether the reason for joint model use affected the proportion of possible MA for each of the longitudinal, time-to-event and association parameter components, we investigated the proportions of possible MA for studies using joint models to account for dropout, or to account for error in a time-varying covariate.
The aim of this review was not to perform any MA, solely to assess if MA were undertaken, what proportion of the identified studies could contribute.
Results
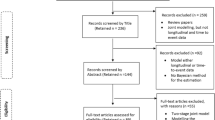
Searches were conducted on the 15th September 2015. The number of references identified is shown in Fig. 1. Once duplicate references were removed, (and an erratum paper correcting an author’s name), 618 references remained. Of these, we identified 210 methodological papers, and disregarded 343 references. In total 65 studies [9–73] remained that applied joint models to data with the aim of influencing healthcare rather than solely presenting new joint modelling methods.
Characteristics of identified studies
Year of publication
The distribution of publication year of the studies was skewed towards more recent dates with median publication year 2014 (interquartile range (IQ) 2011–2014, range 2001–2015). Figure 2 indicates an overall trend (with variation between years) of increasing numbers of applied joint modelling papers published (although the maximum number published in a year was only 20). On this graph we have included lines numbered 1–6 at times when significant joint modelling papers were published. In 1997 Wulfsohn and Tsiatis published a paper commonly cited as one of the first joint modelling papers [74] (line 1). In 2000, Henderson et al [9] extended this methodology, with discussion of different sharing structures between the sub-models (line 2). In 2004 two papers were published, by Tsiatis and Davidian [1] (a review of joint modelling methodology), and by Guo and Carlin [75] (examples of implementation of joint modelling in current software) (line 3). In 2010 Rizopoulos published a paper detailing the R joint modelling package JM [76] (line 4), and 2012 saw the publication of a joint modelling textbook [77], and papers describing joint modelling options in Stata (Crowther et al [78]) and the joineR package in R (Philipson et al [79]) (line 5). Also Crowther et al published further papers on joint modelling in Stata in 2013 [80, 81] (line 6). In addition to these events the number of joint modelling workshops, talks and related conferences has increased in recent years (see https://www.liverpool.ac.uk/translational-medicine/departmentsandgroups/joine-r/workshops/, http://eur.academia.edu/DimitrisRizopoulos/Talks, http://www2.le.ac.uk/departments/health-sciences/research/biostats/staff-pages/mjc76, accessed 28 Nov 2016). Whilst it is unclear which of these publications or events contributed to increases in use of joint modelling methods, an increase is noticeable in the application of joint modelling after 2012.
Full text or abstract
Full articles were obtained for 63 studies (96.9%) [9–21, 23–54, 56–73], with abstracts available for 2 studies (3.1%) [22, 55]. Of the identified studies, some individuals were listed as authors on multiple studies, suggesting that the group of individuals applying joint modelling methods may be limited.
Disease area
The disease areas of the studies were wide ranging (Table 1), with the most common including Cancer, HIV/AIDs, transplant data and cognitive decline. This wide range of disease areas demonstrates the applicability of joint modelling methods to a variety of medical fields, however also indicates that currently finding multiple joint modelling studies applied to the same area to pool in an AD-MA could be problematic.
Journal
The studies identified were published in a range of journals, with 8 journals occurring more than once (Table 1), indicating that there may not currently a preferred journal to present joint modelling studies in.
Reason for use of joint model
The reasons given for using joint models are summarised in Table 1 (with some studies providing multiple reasons giving a total greater than 65). The two most common reasons for joint model use were to investigate the link between the outcomes, or to account for dropout. Only 4 studies stated that they used a joint model to include a time varying covariate in the time-to-event sub-model.
Source of methods used
Of the included studies 18 (27.7%) used study specific modelling methods, whilst the remainder cited methods described in other papers. In total 38 unique papers were cited for methods, with ten cited by more than one study (see Table 2, references included in table). Some papers were software specific (e.g., Rizopoulos 2010 [76] and 2012 [77] are R related, whilst Crowther et al 2013 [81] is Stata related). Others provided methodology and implementation overviews (e.g., Proust-Lima et al [82] and Guo-Carlin [75]).
Modelling approach
Of the 65 included studies, 45 (69.2%) took a frequentist approach, 17 (26.2%) took a Bayesian approach, 1 (1.5%) took both (in separate model fits) and in 2 (3.1%) studies it was unclear (these were the two abstracts). The larger proportion of frequentist approaches may be attributable to the larger number of papers and based on frequentist methods. Additionally the main joint modelling textbook [77] deals with frequentist methods.
There were 21 unique model types recorded for the longitudinal sub-model (plus 1 study with unclear type). Linear mixed effects models were most common (35 studies, 53.8%), followed by mixed effect models with splines (6 studies, 9.2%) or mixed models with unspecified structure (5 studies, 7.7%). Other methods used included different mixed models dependent on latent class, non-linear models with or without splines, and models with change points.
The methods used for the time-to-event sub-model varied widely (and were unclear in 4 studies). The Cox proportional hazards (PH) model was most common (8 studies, 12.3%). Other methods included models with parametric baselines, such as a Weibull PH model (5 studies, 7.7%), a PH model with piecewise constant baseline (4 studies, 6.2%), or a spline modelled baseline hazard (2 studies, 3.1%). Parametric models included the Weibull (5 studies, 7.7%) and the exponential (1 study, 1.5%), and 1 study (1.5%) examined both Weibull and exponential models.
Sharing structure between longitudinal and time-to-event sub-models
The structures used to link the longitudinal and time-to-event sub-models are listed in Table 2. Some studies fitted multiple joint models, with varying sharing structures, allowing a total of more than 65 recorded sharing structures.
Any sharing structure (also termed association structure) involving a function of both fixed and random effects is designated “Fixed and Random effects” whereas those involving random effects but no fixed effects are termed “Random Effects only”. Fixed and random effects sharing structures (33 studies, 50.8%) model effects of aspects of the overall longitudinal outcome value on the time-to-event outcome. The random effects only sharing structures (27 studies, 41.5%) model the effect of individual deviation from the population mean longitudinal outcome on the time-to-event outcome. A description of fixed and random effects sharing structures is given in Rizopoulos 2012 [77], whilst Henderson et al [4] discuss random effects only sharing structures. Additionally Rizopoulos and Ghosh [83] and Gould [84] discuss a range of association structures.
The fixed and random effect sharing structures can be subdivided further. Current value refers to models inserting the current longitudinal trajectory value into the time-to-event sub-models, and is used when the current overall value of the longitudinal trajectory affects the risk of an event. The current slope or first derivative of the population trajectory could also be inserted into the time-to-event sub-model, in conjunction with the current value or alone, and is used to model the effect of rate of change of the longitudinal variable on the risk of an event. Another fixed and random effects sharing structure identified in one study, inserted the fixed and random effects coefficients of the longitudinal trajectory into the time-to-event sub-model without their related covariates.
Random effects only sharing structures can also be grouped. We define a random effects only sharing structure to contain covariates if it is of format such as α(U 0 + U 1 t) or α 1 U 0 + α 2 U 1 + α 3(U 0 + U 1 t) where the α terms are association parameters, the U terms are random effects, and t represents a covariate such as time. Alternatively, if the structure is similar to α(U 0 + U 1), where the random effect U 1 had a covariate t in the longitudinal sub-model, we define the random effects only sharing structure to not contain covariates (see Henderson et al [9] for further examples).
The specialist sharing structure group (4 studies (6.2%)) contained less common sharing structures such as associating the time-to-event and longitudinal sub-models through a multivariate distribution. Another option was the latent class structure [3, 82], used in at least one joint model in 3 studies (4.6%). Finally 4 (6.2%) studies, including the 2 abstracts, had unclear sharing structures.
We should note that choice of association structure should be driven by the data itself, and so it is expected to see a range of sharing structures given the range of disease areas of the identified studies.
Software
The software and package used in the included studies is listed in Table 3. Packages/methods have been stated in Table 3 even if no identified studies currently used them.
Software used was not always stated by identified studies, a potential issue for future MA when determining exact modelling structure used. The most mentioned software was R, although SAS and Stata were also common. Some of the software identified requires more coding from users (such as C++ WinBUGs), which might explain the preference for software with specific joint modelling packages.
The current preference in R is for the JM package [76] (implements frequentist joint models that insert the fixed and random effects of the longitudinal sub-model into the time-to-event sub-model), and PROC NLMIXED in SAS (allows fitting of a range of non-linear mixed models). The popularity of JM might be explained by the availability of a textbook with worked examples of joint model implementation using the package [77].
In four studies (6.2%) more than one software was stated, it was unclear which implemented the joint model fit.
Did studies report sufficient information to contribute to meta-analyses?
For an identified study to contain sufficient information to contribute to a MA, it must report a sample size. For each meta-analysis group (longitudinal, time-to-event, association), the relevant parameter estimates must be reported with a precision estimate (a standard error, or a confidence interval with related significance level). A summary of this information is given in Table 4.
The sample size was reported in the majority (98.5%) of the identified studies, with median sample size of 514 (IQR 277–1054.5, range 46–3814).
The association parameters were more commonly reported (51 studies (78.5%)) than those of the longitudinal and time-to-event sub-models (45 (69.2%) and 46 (70.8%) respectively). This could be attributable to the high proportion of identified studies that stated that joint models were used to investigate the link between longitudinal and time-to-event outcomes.
The number of studies where a precision measure was available was comparable to the number of studies that reported coefficients, across the three MA categories. Whilst a MA would be possible for each category if the parameter, the standard error and the sample size were reported, if the only available precision estimate was the confidence interval then the significance level was also required. In the studies we identified, the significance level was unclear for 8 (12.3%) studies, was 0.05 for 53 (81.5%) studies, 0.01 for 3 (4.6%) studies, 0.1 for 1 (1.5%) study.
Overall, a MA would be possible for the association parameter in 50 (76.9%) studies, for the longitudinal parameters in 44 (67.7%) studies, and for time-to-event parameters in 45 (69.2%) studies.
Ideally, a study would provide enough information to perform MA in all three separate groups. Sufficient information to allow all three MA to be undertaken was available from 38 (58.5%) of the studies. Only two MA were possible in 6 studies (9.2%), only 1 in 13 studies (20.0%) and in 8 studies (12.3%) there was not sufficient information to complete any MA.
The reasons for joint model use may affect the information stated in the study report. We re-examined the proportions of studies in which the three groups of MA could take place, dependent on the reason for joint model use (Table 4).
For the 22 studies which stated accounting for dropout as a reason for joint model use, we saw a much higher percentage (81.8%) for which MA of longitudinal sub-model parameters was possible, compared to for all studies (67.7%). However percentages of MA possible for the time-to-event coefficients or association parameters were smaller. This could be explained by studies using joint models to account for dropout being mainly interested in the parameters from the longitudinal sub-model.
Only 4 studies stated inclusion of a time varying covariate in a time-to-event model as one of their reasons for using joint models. There was a slight indication that the longitudinal coefficients for studies using joint models to include time varying covariates in time-to-event models are worse reported than for all identified studies, possibly because the longitudinal component of the joint model is of interest as a covariate rather than an outcome in these cases, however more information is needed before this relationship can be fully investigated.
Discussion
Joint models for longitudinal and time-to-event data are often stated as beneficial compared to separate longitudinal or time-to-event analyses, as they can reduce bias and increase efficiency in model estimation (see Ibrahim et al 2010 [85] for example). Additionally Powney et al 2014 [86] discuss a study where joint models showed a significant difference between treatment groups that was not identified by separate analyses [87]. These benefits of joint models reinforce the suggestion that in certain circumstances MA of joint models may be more appropriate than MA of separate models.
We aimed through our search strategy to identify all studies that implemented joint models to influence future healthcare, and believe that the studies identified are representative of the current literature. However if studies did not state the key search terms used in this review (see Additionnal file S1 for search strategies) in text accessible to the search, the study may not have been identified. For example, from Powney et al [86] we know that the MAGNETIC trial [87] utilised joint models, however this is not mentioned in the abstract. When joint models are not used as part of the primary analyses, their use may be unclear from the abstract or keywords. Therefore we include statement of statistical methods used in text accessible to search engines to our recommendations for future reporting of joint models, stated in Table 5.
With the increasing use of joint models in the literature, ensuring they are well reported is vital so that the analyses can be interpreted fully and that the published data can be used in future evidence synthesis. We have identified that for the scenario of AD-MA of the results of joint models published in the literature, it was possible to perform MA from a high proportion but not all studies. We would recommend for future practice that regardless of the reason for joint model use, full model covariates with precision estimates be reported either in the study report or supplementary materials (Table 5) not only to aid interpretation within the study itself, but to ensure that future MA would be possible.
Additionally it is important that model structure (both sub-models and the sharing structures that link them) are clearly reported (Table 5). However in some cases model formulae were not reported, a particular issue identified for association parameters. We have noted the multiple association structures available to researchers. Due to the different interpretation of each, it may not make sense to pool association parameters from radically different sharing structures. Without clear statement of the model structure it may be difficult to conduct a future MA. It would be beneficial if studies applying joint models included statement of the model structure as standard to give clarity to the methods used.
Also, it is important especially in joint modelling to state the software and packages or functions used, as the software used could indicate the model structure, as well as methods used to obtain the parameter estimations (Table 5).
Consistently, the proportion of studies where association parameter MA was possible was higher than the proportion where MA was possible for longitudinal or time-to-event parameters. The association information may have been more commonly reported than the longitudinal or time-to-event information because the association parameters in shared random effect models quantify the link between the sub-models. It can be expected therefore that studies aiming to quantify the link between the sub-models report the association information more prominently than other model parameters.
Conclusion
Overall, this investigation has highlighted the need to fully report the coefficients and precision estimates in studies applying joint models to datasets. Whilst this review identified a limited number of studies that fulfilled our criteria, the range of disease areas covered by the studies was large with applications using a wide range of sub-models and association structures being published in an assortment of journals by a range of authors. It may be some years before there are sufficient studies published in one area to conduct a meta-analysis of joint models. Nevertheless our review has demonstrated that the use of joint models is increasing every year and the availability of software to fit a range of flexible models is likely to facilitate their application even further.
In the future the current standard of joint modelling should be maintained and improved upon, following the recommendations given in this paper, in order that information from published studies can be used for the purpose of MA.
Change history
04 April 2018
Following publication of the original article [1] the authors reported that reference 15 (Cella et al.) had been incorrectly replaced with a duplicate of Brombin et al. during publication.
References
Tsiatis AA, Davidian M. Joint modeling of longitudinal and time-to-event data: An overview. Stat Sinica. 2004;14(3):809–34.
Gould AL, et al. Responses to discussants of 'Joint modeling of survival and longitudinal non-survival data: Current methods and issues. report of the DIA Bayesian joint modeling working group'. Stat Med. 2015;34(14):2202–3.
Proust-Lima C, et al. Joint latent class models for longitudinal and time-to-event data: A review. Stat Methods Med Res. 2014;23(1):74–90.
Henderson R, Diggle P, Dobson A. Joint modelling of longitudinal measurements and event time data. Biostatistics. 2000;1(4):465–80.
Glass GV. Primary, Secondary, and Meta-Analysis of Research. Educ Res. 1976;5(10):3–8.
Higgins J, Green S. and (editors). Cochrane Handbook for Systematic Reviewsof Interventions Version 5.1.0 (updated March 2011). The Cochrane Collaboration, 2011. Available from http://handbook.cochrane.org/.
Whitehead A. Meta-analysis of controlled clinical trials/Anne Whitehead. Statistics in practice. Chichester: John Wiley & Sons; 2002.
Moher D, et al. Methods of Systematic Reviews and Meta-Analysis: Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. J Clin Epidemiol. 2009;62:1006–12.
Andrinopoulou ER, et al. An introduction to mixed models and joint modeling: Analysis of valve function over time. Ann Thorac Surg. 2012;93(6):1765–72.
Andrinopoulou ER, et al. Joint modeling of two longitudinal outcomes and competing risk data. Stat Med. 2014;33(18):3167–78.
Argyropoulos C, et al. Dialyzer reuse and outcomes of high flux dialysis. PLoS ONE. 2015;10(6):e0129575.
Artaud F, et al. Decline in fast gait speed as a predictor of disability in older adults. J Am Geriatr Soc. 2015;63(6):1129–36.
Berzuini C, Allemani C. Effectiveness of potent antiretroviral therapy on progression of human immunodeficiency virus: Bayesian modelling and model checking via counterfactual replicates. J Royal Stat Soc Series C Appl Stat. 2004;53(4):633–50.
Brombin C, Di Serio C, Rancoita PM. Joint modeling of HIV data in multicenter observational studies: A comparison among different approaches. Stat Methods Med Res. 2016;25(6):2472–87. Epub 2014 Mar 26.
Brombin C, Di Serio C, Rancoita PM. Joint modeling of HIV data in multicenter observational studies: A comparison among different approaches. Stat Methods Med Res. 2016;25(6):2472-487. Epub 2014Mar 26.
Chang CCH, et al. Minimizing attrition bias: a longitudinal study of depressive symptoms in an elderly cohort. Int Psychogeriatr. 2009;21(5):869–78.
Chen Q, et al. Joint modeling of longitudinal and survival data with missing and left-censored time-varying covariates. Stat Med. 2014;33(26):4560–76.
Daher Abdi Z, et al. Impact of longitudinal exposure to mycophenolic acid on acute rejection in renal-transplant recipients using a joint modeling approach. Pharmacol Res. 2013;72:52–60.
Dantan E, et al. Joint model with latent state for longitudinal and multistate data. Biostatistics. 2011;12(4):723–36.
Deslandes E, Chevret S. Joint modeling of multivariate longitudinal data and the dropout process in a competing risk setting: Application to ICU data. BMC Med Res Methodol. 2010;10:69.
Du H, Hahn EA, Cella D. The impact of missing data on estimation of health-related quality of life outcomes: An analysis of a randomized longitudinal clinical trial. Health Services and Outcomes Research Methodology. 2011;11(3-4):134–44.
Duvignac J, et al. CD4+ T-lymphocytes natural decrease in HAART-naive HIV-infected adults in Abidjan. HIV Clinical Trials. 2008;9(1):26–35.
Ediebah DE, et al. Joint modeling of longitudinal health-related quality of life data and survival. Qual Life Res. 2015;24(4):795–804.
Elhaj M, et al. Can serum surfactant protein D or CC-chemokine ligand 18 predict outcome of interstitial lung disease in patients with early systemic sclerosis? J Rheumatol. 2013;40(7):1114–20.
Fairclough DL, et al. Handling missing quality of life data in HIV clinical trials: what is practical? Qual Life Res. 2008;17(1):61–73.
Garre FG, et al. A joint latent class changepoint model to improve the prediction of time to graft failure. J Royal Stat Soc Series A Stat Soc. 2008;171(1):299–308.
Ghisletta P. Application of a joint multivariate longitudinal-survival analysis to examine the terminal decline hypothesis in the Swiss Interdisciplinary Longitudinal Study on the Oldest Old. J Gerontol B Psychol Sci Soc Sci. 2008;63(3):185–92.
Ghisletta P, McArdle JJ, Lindenberger U. Longitudinal cognition-survival relations in old and very old age: 13-year data from the Berlin aging study. Eur Psychol. 2006;11(3):204–23.
Goyal N, Gomeni R. A latent variable approach in simultaneous modeling of longitudinal and dropout data in schizophrenia trials. Eur Neuropsychopharmacol. 2013;23(11):1570–6.
Graham PL, Ryan LM, Luszcz MA. Joint modelling of survival and cognitive decline in the Australian Longitudinal Study of Ageing. J Royal Stat Soc Series C Appl Stat. 2011;60(2):221–38.
Guler I, et al. Joint modelling for longitudinal and time-to-event data: Application to liver transplantation data, in 14th International Conference on Computational Science and Its Applications, ICCSA 2014. Guimaraes: Springer Verlag; 2014. p. 580–93.
Husser O, et al. Tumor marker carbohydrate antigen 125 predicts adverse outcome after transcatheter aortic valve implantation. JACC Cardiovasc Interv. 2013;6(5):487–96.
Ibrahim JG, Chen MH, Sinha D. Bayesian methods for joint modeling of longitudinal and survival data with applications to cancer vaccine trials. Stat Sin. 2004;14(3):863–83.
Jacoby A, et al. Quality-of-life outcomes of initiating treatment with standard and newer antiepileptic drugs in adults with new-onset epilepsy: findings from the SANAD trial. Epilepsia. 2015;56(3):460–72.
Khoundabi B, et al. Acute kidney injury in ICU patients following non-cardiac surgery at Masih Daneshvari hospital: joint modeling application. Tanaffos. 2015;14(1):49–54.
Kupferman JC, et al. BP control and left ventricular hypertrophy regression in children with CKD. J Am Soc Nephrol. 2014;25(1):167–74.
Kypriotakis G, Vidrine DJ, Francis LE, Rose JH. The longitudinal relationship between quality of life and survival in advanced stage cancer. Psychooncology. 2016;25(2):225–31. doi:10.1002/pon.3846. Epub 2015 May 8.
Lawson A.B, et al. Bayesian latent structure modeling of walking behavior in a physical activity intervention. Stat Methods Med Res. 2014
Levine SZ, et al. Joint modeling of dropout and outcome in three pivotal clinical trials of schizophrenia. Schizophr Res. 2015;164(1-3):122–6.
Liu X, et al. Does C-reactive protein predict the long-term progression of interstitial lung disease and survival in patients with early systemic sclerosis? Arthritis Care and Research. 2013;65(8):1375–80.
Malehi AS, et al. Assessing the autoantibody levels in relation to recurrence of pemphigus: Joint modeling of longitudinal measurements and recurrent event times. Iran Red Crescent Med J. 2014;16(2):e13812.
Marioni RE, et al. Cognitive lifestyle jointly predicts longitudinal cognitive decline and mortality risk. Eur J Epidemiol. 2014;29(3):211–9.
Mayeda ER1, Haan MN, Yaffe K, Kanaya AM, Neuhaus J. Does type 2 diabetes increase rate of cognitive decline in older Mexican Americans? Alzheimer Dis Assoc Disord. 2015;29(3):206-12. doi:10.1097/WAD.0000000000000083.
McLain AC, Sundaram R, Buck Louis GM. Joint analysis of longitudinal and survival data measured on nested timescales by using shared parameter models: An application to fecundity data. J Royal Stat Soc Series C Appl Stat. 2015;64(2):339–57.
Medeiros FA, et al. Evaluation of progressive neuroretinal rim loss as a surrogate end point for development of visual field loss in glaucoma. Ophthalmology. 2014;121(1):100–9.
Meira-Freitas D, et al. Predicting progression in glaucoma suspects with longitudinal estimates of retinal ganglion cell counts. Investig Ophthalmol Vis Sci. 2013;54(6):4174–83.
Meira-Freitas D, et al. Predicting progression of glaucoma from rates of frequency doubling technology perimetry change. Ophthalmology. 2014;121(2):498–507.
Miki A, et al. Rates of retinal nerve fiber layer thinning in glaucoma suspect eyes. Ophthalmology. 2014;121(7):1350–8.
Murphy TE, et al. A longitudinal, observational study with many repeated measures demonstrated improved precision of individual survival curves using bayesian joint modeling of disability and survival. Exp Aging Res. 2015;41(3):221–39.
Njagi EN, et al. A joint survival-longitudinal modelling approach for the dynamic prediction of rehospitalization in telemonitored chronic heart failure patients. Stat Model. 2013;13(3):179–98.
Núñez J, et al. Red blood cell distribution width is longitudinally associated with mortality and anemia in heart failure patients. Circ J. 2014;78(2):410–8.
Pan J, et al. Joint longitudinal and survival-cure models in tumour xenograft experiments. Stat Med. 2014;33(18):3229–40.
Pauler DK, Finkelstein DM. Predicting time to prostate cancer recurrence based on joint models for non-linear longitudinal biomarkers and event time outcomes. Stat Med. 2002;21(24):3897–911.
Paulsen JS, et al. Prediction of manifest Huntington's disease with clinical and imaging measures: a prospective observational study. Lancet Neurol. 2014;13(12):1193–201.
Pike F, Weissfeld LA, Chang CCH. Joint modeling of multivariate censored longitudinal and event time data with application to the Genetic Markers of Inflammation Study. J Appl Stat. 2014;41(10):2178–91.
Pilla Reddy V, et al. Modelling and simulation of the positive and negative syndrome scale (PANSS) time course and dropout hazard in placebo Arms of schizophrenia clinical trials. Clin Pharmacokinet. 2012;51(4):261–75.
Sargent-Cox KA, Anstey KJ, Luszcz MA. Longitudinal change of self-perceptions of aging and mortality. J Gerontol B Psychol Sci Soc Sci. 2014;69(2):168–73.
Seid A, et al. Joint modeling of longitudinal CD4 cell counts and time-to-default from HAART treatment: A comparison of separate and joint models. Electron J Appl Stat Anal. 2014;7(2):292–314.
Shen Y, et al. Joint modeling tumor burden and time to event data in oncology trials. Pharm Stat. 2014;13(5):286–93.
Skupien J, et al. Synergism between circulating tumor necrosis factor receptor 2 and HbA(1c) in determining renal decline during 5-18 years of follow-up in patients with type 1 diabetes and proteinuria. Diabetes Care. 2014;37(9):2601–8.
Su CT, et al. Changes in anthropometry and mortality in maintenance hemodialysis patients in the HEMO study. Am J Kidney Dis. 2013;62(6):1141–50.
Tang X, et al. Increasing Chimerism after Allogeneic Stem Cell Transplantation Is Associated with Longer Survival Time. Biol Blood Marrow Transplant. 2014;20(8):1139–44.
Terrin N, Rodday AM, Parsons SK. Joint models for predicting transplant-related mortality from quality of life data. Qual Life Res. 2015;24(1):31–9.
Thabut G, et al. Survival benefit of lung transplant for cystic fibrosis since lung allocation score implementation. Am J Respir Crit Care Med. 2013;187(12):1335–40.
Thiebaut R, et al. Joint modelling of bivariate longitudinal data with informative dropout and left-censoring, with application to the evolution of CD4+ cell count and HIV RNA viral load in response to treatment of HIV infection. Stat Med. 2005;24(1):65–82.
van den Hout A, Muniz-Terrera G. Joint models for discrete longitudinal outcomes in aging research. J. R. Stat. Soc. C. 2016;65:167–186. doi:10.1111/rssc.12114 Issue online: 17 December 2015.
Van Der Linde D, et al. Natural history of discrete subaortic stenosis in adults: A multicentre study. Eur Heart J. 2013;34(21):1548–56.
Wang Y, Taylor JMG. Jointly Modeling Longitudinal and Event Time Data with Application to Acquired Immunodeficiency Syndrome. J Am Stat Assoc. 2001;96(455):895–905.
Watanabe H, et al. Factors affecting longitudinal functional decline and survival in amyotrophic lateral sclerosis patients. Amyotroph Lateral Scler Frontotemporal Degeneration. 2015;16(3-4):230–6.
Wu L, Liu W, Hu XJ. Joint inference on HIV viral dynamics and immune suppression in presence of measurement errors. Biometrics. 2010;66(2):327–35.
Xue QL, et al. Heterogeneity in rate of decline in grip, hip, and knee strength and the risk of all-cause mortality: The women's health and aging study II. J Am Geriatr Soc. 2010;58(11):2076–84.
Yu M, Taylor JMG, Sandler HM. Individual prediction in prostate cancer studies using a joint longitudinal survival-cure model. J Am Stat Assoc. 2008;103(481):178–87.
Zhang JP, et al. Joint modeling of longitudinal changes in depressive symptoms and mortality in a sample of community-dwelling elderly people. Psychosom Med. 2009;71(7):704–14.
Wulfsohn MS, Tsiatis AA. A joint model for survival and longitudinal data measured with error. Biometrics. 1997;53(1):330–9.
Guo X, Carlin BP. Separate and Joint Modeling of Longitudinal and Event Time Data Using Standard Computer Packages. Am Stat. 2004;58(1):16–24.
Rizopoulos D. JM: An R Package for the Joint Modelling of Longitudinal and Time-to-Event Data. J Stat Softw. 2010;35(9):1–33.
Rizopoulos D. Joint models for longitudinal and time-to-event data, with applications in R. Boca Raton: Chapman and Hall/CRC; 2012.
Crowther MJ, Abrams KR, Lambert PC. Flexible parametric joint modelling of longitudinal and survival data. Stat Med. 2012;31(30):4456–71.
Philipson, P, et al. joineR: Joint modelling of repeated measurements and time-to-event data. Comprehensive R Archive Network, 2012.
Crowther, Michael J., (2013), STJM: Stata module to fit shared parameter joint models of longitudinal and survival data, http://EconPapers.repec.org/RePEc:boc:bocode:s457502.
Crowther MJ, Abrams KR, Lambert PC. Joint modeling of longitudinal and survival data. Stata J. 2013;13(1):165–84.
Proust-Lima C, et al. Joint modelling of multivariate longitudinal outcomes and a time-to-event: A nonlinear latent class approach. Comput Stat Data Anal. 2009;53(4):1142–54.
Rizopoulos D, Ghosh P. A Bayesian semiparametric multivariate joint model for multiple longitudinal outcomes and a time-to-event. Stat Med. 2011;30(12):1366–80. 15.
Lawrence Gould A, et al. Joint modeling of survival and longitudinal non-survival data: Current methods and issues. Report of the DIA Bayesian joint modeling working group. Stat Med. 2015;34(14):2181–95.
Ibrahim JG, Chu H, Chen LM. Basic concepts and methods for joint models of longitudinal and survival data. J Clin Oncol. 2010;28(16):2796–801.
Powney M, et al. A review of the handling of missing longitudinal outcome data in clinical trials. Trials. 2014;15.
Powell C, et al. MAGNEsium Trial In Children (MAGNETIC): a randomised, placebo-controlled trial and economic evaluation of nebulised magnesium sulphate in acute severe asthma in children. Health Technol Assess. 2013;17(32):1–216. 216p.
Wulfsohn MS, Tsiatis AA. A joint model for survival and longitudinal data measured with error. Int Biometric Soc. 1997;53(1):330–9.
Diggle PJ, Sousa I, Chetwynd AG. Joint modelling of repeated measurements and time-to-event outcomes: The fourth Armitage lecture. Stat Med. 2008;27(16):2981–98.
Rizopoulos D. Dynamic Predictions and Prospective Accuracy in Joint Models for Longitudinal and Time-to-Event Data. Biometrics. 2011;67(3):819–29.
R Core Team and R: A language and environment for statistical computing. In R Foundation for Statistical Computing, Vienna, Austria. 2015. https://www.R-project.org/.
Rizopoulos D. The R Package JMbayes for Fitting Joint Models for Longitudinal and Time-to-Event Data using MCMC. 2014
Rondeau V, Mazroui Y, Gonzalez JR. Frailtypack: An r package for the analysis of correlated survival data with frailty models using penalized likelihood estimation or parametrical estimation. J Stat Softw. 2012;47.
SAS software. Copyright, SAS Institute Inc. SAS and all other SAS Institute Inc. product or service names are registered trademarks or trademarks of SAS Institute Inc., Cary, NC, USA.
Garcia-Hernandez, A. and Rizopoulos, D. (2016). %JM: A SAS macro to fit jointly generalized mixed models for longitudinal data and time-to-event responses. Journal of Statistical Software, to appear.
Zhang D, et al. JMFit: A SAS Macro for Assessing Model Fit in Joint Models of Longitudinal and Survival Data. J Stat Softw. 2015. in press.
StataCorp. 2015. Stata Statistical Software: Release 14. College Station, TX: StataCorp LP.
Lunn DJ, et al. WinBUGS - A Bayesian modelling framework: Concepts, structure, and extensibility. Stat Comput. 2000;10(4):325–37.
Beal SL, S. L.B. NONMEM Users Guides. Icon Development Solutions. Ellicott City, Maryland, USA. 1989-98.
ISO International Standard ISO/IEC 14882:2014(E) – Programming Language C++.
Muthén L.K and Muthén B.O. Mplus User’s Guide. Sixth Edition. Los Angeles, CA: Muthén & Muthén; 1998-2010.
Acknowledgements
Not applicable.
Funding
This work was supported by the Health eResearch Centre (HeRC) funded by the Medical Research Council Grant MR/K006665/1.
Availability of data and material
The datasets analysed during the review are available from the corresponding author on reasonable request. The data collection form is included in the Additional file 2.
Authors’ contributions
MS, RKD and CTS all participated in discussion of the design of the investigation. MS extracted data from the identified studies, conducted analyses and was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Competing interests
MS and CTS were authors on one of the studies identified in this review. RKD was involved in the original grant for the development of the R joint modelling package joineR.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1:
This file includes the search strategies used in this review to search Medline, Pubmed and Scopus. (DOCX 13 kb)
Additional file 2:
This file includes a blank example of the data collection form used to record information from the studies identified by this review. (DOCX 14 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Sudell, M., Kolamunnage-Dona, R. & Tudur-Smith, C. Joint models for longitudinal and time-to-event data: a review of reporting quality with a view to meta-analysis. BMC Med Res Methodol 16, 168 (2016). https://doi.org/10.1186/s12874-016-0272-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12874-016-0272-6