Abstract
Background
Our study aims to explore the association of rs7025486 single-nucleotide polymorphisms (SNP) in DAB2IP and rs1333049 on chromosome 9p21.3 with the coronary artery disease in Chinese population.
Methods
All patients came from the east China area and underwent coronary angiography. Rs7025486 and rs1333049 polymorphism were genotyped in 555 patients with CAD and in 480 healthy controls that underwent coronary angiography.
Results
In Chinese population, the rs7025486 genotype in the case group was no significant different than the control group (P = 0.531).Meanwhile, the rs1333049 SNP has statistically significant (P = 0.006), which was the independent risk factors for CAD (OR1.252, P = 0.039), and consistent with the past studies conclusion.
Conclusion
Genotype of rs1333049 on chromosome 9p21, but not rs7025486 on chromosome 9q33, is an independent determinant of the incidence of CAD in Chinese population.
Similar content being viewed by others

Background
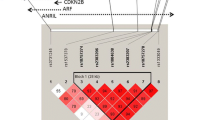
Coronary artery disease (CAD) is a chronic multifactor inflammatory disease which can progress to acute coronary syndrome and sudden cardiac death [1]. CAD is associated with a family history as well as several established risk factors including diabetes mellitus, hypertension, hyperlipidemia and smoking, suggesting that the pathogenesis of CAD has a substantial genetic component [2]. Genome-wide association studies (GWAS) have identified several genetic variants that increased susceptibility to CAD and acute myocardial infarction (AMI) in the primary prevention setting [3]. The strongest association signal in the genome in GWAS studies for CAD and AMI that has been published thus far comes from a number of single-nucleotide polymorphisms (SNPs) with a high degree of linkage disequilibrium between each individual on chromosome 9p21 [4,5,6]. However, several subsequent studies have shown inconsistent results when the association between these genetic variants and CAD or AMI was examined [7, 8]. In addition, these data are mostly from American African, Caucasian, South Korean, and Japanese cohorts. Most of SNPs on 9p21 are located within a long non-coding RNA, namely antisense non-coding RNA in the INK4 locus (ANRIL). It seems plausible that the influence of ANRIL on CAD is mediated by the upstream genes CDKN2A and CDKN2B. The dysfunction of CDKN2A and CDKN2B subsequently causes excessive cell proliferation [9,10,11].
Recently, a European GWAS reported that chromosome 9q33 contains a novel susceptibility locus DAB2IP associated with abdominal aortic aneurysm, early onset myocardial infarction, peripheral artery disease and pulmonary embolism [12]. Nevertheless, relevant reports on the association of rs7025486 located within 9q33 with CAD were lacking in Chinese population. DAB2IP is considered as a Ras-GTPase activator and a tumor suppressor gene, repressing tumor proliferation and metastasis and maintaining chromosomal stability [13, 14].
Toward this end, we selected two SNPs, which represented the most associated and independent SNP rs1333049 at 9p21.3 and rs7025486 at 9q33 in the previous studies, to investigate their role in predicting CAD.
Methods
Study population
The study protocol was approved by the hospital ethics committee, and written informed consents were obtained from all subjects. The study population consisted of 1151 Chinese Han patients undergoing coronary angiography to evaluate suspected or established CAD. Sixty-four patients with type 1 diabetes mellitus were identified by measuring C peptide levels and excluded; we also excluded 52 patients with chronic viral or bacterial infections, tumors, or immune system disorders. Of the final 1035 enrollments, 555 patients had significant CAD (≥50% luminal diameter narrowing in at least one coronary artery), and 480 were considered to be healthy controls [15]. Type 2 diabetes mellitus was referred to as a fasting plasma glucose level of ≥7.0 mmol/L or a non-fasting plasma glucose level of ≥11.1 mmol/L, or taking oral hypoglycemic drugs or receiving parenteral insulin therapy [15]. Patients were diagnosed with hyperlipidemia if they had serum levels of total cholesterol (TC) >5.7 mmol/L (220 mg/dl), low-density lipoprotein cholesterol (LDL-C) >3.64 mmol/L (140 mg/dl), triglycerides (TG) >1.7 mmol/L (150 mg/dl) or high-density lipoprotein cholesterol (HDL-C) <0.91 mmol/L (35 mg/dl). Early-onset CAD was considered as clinical CAD occurring by age ≤ 55 years in male or ≤60 years in female patients [9].
Genotype determination
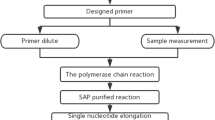
Genomic DNA was extracted from peripheral blood cells according to the manufacturer’s instrument (Tiagen Biotech, China) [15]. Genotyping was performed with TaqMan SNP allelic discrimination by means of an ABI 7900HT (Applied Biosystems, Foster City, CA, USA), in a 384-well format [16]. The TaqMan Assay kit was obtained from Applied Biosystems (Foster City, USA). Genotypes were determined as previously described [16]. Finally, we applied the ABI Prism SDS software version 2.1 to analyze the data.
Statistical analysis
Continuous variables are expressed as mean ± standard deviation, while categorical data are presented as frequencies or percentages. The differences among groups were determined by Student t test or one-way ANOVA analysis [16]. The differences in categorical data among groups were determined by the chi-square test. Odds ratios (ORs) of CAD for CC genotype on rs1333049, and other risk factors, were estimated by multivariate Logistic regression analyses [17]. A 2-sided probability level of ≤0.05 was considered significant. All analyses were performed with SPSS for Windows 13.0 (SPSS Inc., Chicago, Illinois, USA).
Results
Baseline characteristics of enrollments
The baseline characteristics of the study groups are presented in Table 1, which show the expected differences in classical risk factors in CAD patients compared with controls. CAD patients in our studies are predominance of male subjects (62.52%). Furthermore, patients with CAD have higher incidence rate of smoking and diabetes mellitus compared with healthy controls (43.96% vs. 20.42% and 67.39% vs. 31.25%, respectively). There are no differences in age, history of hypertension and hyperlipidemia, family history of CAD, and levels of blood urine nitrogen (BUN) and uric acid between two groups.
Distribution of rs1333049 and rs7025486 genotype between CAD patients and controls
The observed rs1333049 and rs7025486 genotype frequencies did not deviate from Hardy-Weinberg equilibrium (Table 1, P > 0.05 using a chi-squared goodness-of-fit model). This indicated that the case and control groups were representative of the population and had no selection bias. Table 2 shows the distribution of rs1333049 and rs7025486 genotype in our study. The distribution of rs1333049 genotypes significantly differed between CAD patients and healthy controls (P = 0.006). In contrast, no discrepancy was found in the distribution of rs7025486 genotype between two groups (P = 0.531). The whole enrollments were then divided into several subgroups according to the presence of history of diabetes mellitus, hypertension, AMI and early onset myocardial infarction. However, we could not find a positive association of rs7025486 genotype with these conditions (Table 3).
Rs1333049 is an independent determinant of the incidence of CAD
To gain further insight into the role of rs1333049 in independently predicting CAD, multivariate logistic regression was performed to further analyze the data by adjusting for gender, age, history of smoking, diabetes mellitus, fasting glucose, hypertension and hyperlipidemia. As shown in Table 4, apart from established risk factors including smoking, diabetes mellitus and hypertension, rs1333049 remained associated with the incidence of CAD, suggesting that rs1333049 genotype was an independent determinant of the incidence of CAD.
Discussion
Since the fact that CAD and other vascular diseases share common risk factors, including genetic variants and environmental risk factors associated with peripheral vascular disease are expected candidates affecting the risk of CAD [18, 19]. In our case–control study, two previously reported SNPs representing the genetic variants on 9p21 and 9q33 were chosen to investigate the association with CAD. Our study validated that rs1333049 at chromosome 9p21 showed a significant association with CAD, whereas variant at 9q33 showed no association with CAD and main cardiovascular risk factors in our data.
Accumulating evidence came to conclusion that the incidence of hypertension and diabetes mellitus, higher inflammatory response and LDL levels are the risk factors for atherosclerotic progression [20]. Furthermore, based on optical coherence tomography, cholesterol, hs-CRP and pentraxin 3 were associated with thin-cap fibroatherma, which is known as vulnerable plaques [20,21,22]. Similarly, some of these known risk factors were referred to as independent determinants of CAD in our study.
In the past decades, a number of novel susceptibility genes of CAD were identified using GWASs [19]. In particular, several SNPs on chromosome 9p21 identified by the Welcome Trust Case Control Cohort study (WTCCC), McPherson et al. [4] and Helgadottir et al. [3] met the criteria for genome-wide association. Consistent with these results, SNP rs1333049, which has been reported in other Asian population, is strongly associated with CAD in our case-control study and could be regarded confirmatory [23,24,25]. Based on multivariate analysis, we further collaborated that rs1333049 genotype is not secondary drift to other cardiovascular risk factors and an independent determinant of the incidence of CAD.
A mechanism behind the link between risk alleles on chromosome 9p21 and cardiovascular disease is actively being explored. Recent studies have reported that the expression of the upstream genes CDKN2A and CDKN2B as well as the long non-coding ANRIL was considered to be linked with the risk genotype [9, 10]. None of the SNPs in the haplotype block harbor in the transcribed regions, and thus a change of expression level ascribed to alteration of a promoter or enhancer region is indeed a plausible hypothesis. On the other hand, targeted depletion of the non-coding interval on human chromosome 9p21 in mice provided direct evidence that the risk interval has an important role in regulation of cardiac CDKN2A and CDKN2B expression, suggesting that the region modulates CAD progression by altering the dynamics of vascular cell proliferation [11].
On the other hand, no association between variant in rs7025486 and the incidence of CAD was observed, while the GWAS and replicating study consistently found the effects of a common variant in DAB2IP (rs7025486) on the development of CAD and other complications [12, 26, 27]. In cancer cells, DAB2IP, as a Ras-GTPase, exerted a suppressive effect on tumor invasion and maintained chromosomal stability [13, 14, 28]. However, the molecular mechanisms of DAB2IP in regulating the progression of atherosclerosis need further investigation. Leading explanations for these discrepant findings include the presence of different ethnic groups as well as relative small sample size. In fact, the frequencies of the risk-association alleles in chromosome 9q33 are similar in American African and Caucasian populations, but substantially lower in Asian descent. Thus, failing to identify any significant association of rs7025486 with the incidence of CAD in Asian populations could be attributed to substantially lower statistical power caused by the relatively lower prevalence of the risk allele. In addition, study design or small sample size may also affect the results. This is a hospital-based study in nature. In this regard, the participants in control group may not absolutely healthy and the overestimation of the proportion of healthy participants could result in selection bias in our study.
Conclusions
Genotype of rs1333049 on chromosome 9p21, but not rs7025486 on chromosome 9q33, is an independent determinant of the incidence of CAD in Chinese population.
Abbreviations
- AMI:
-
Acute myocardial infarction
- ANRIL:
-
Antisense non-coding RNA in the INK4 locus
- CAD:
-
Coronary artery disease
- CDKN2A/2B:
-
Cyclin dependent kinase inhibitor 2A/2B
- DAB2IP:
-
DAB2 interacting protein
- GWAS:
-
Genome-wide association studies
- ORs:
-
Odds ratios
- SNPs:
-
Single-nucleotide polymorphisms
- WTCCC:
-
Welcome trust case control cohort study
References
Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352(16):1685–95.
Humphries SE, Drenos F, Ken-Dror G, Talmud PJ. Coronary heart disease risk prediction in the era of genome-wide association studies: current status and what the future holds. Circulation. 2010;121(20):2235–48.
Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–3.
McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–91.
Welcome T. Case, control, consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78.
Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357(5):443–53.
Schunkert H, Gotz A, Braund P, McGinnis R, Tregouet DA, Mangino M, et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117(13):1675–84.
Xie F, Chu X, Wu H, Sun W, Shen M, Yang L, et al. Replication of putative susceptibility loci from genome-wide association studies associated with coronary atherosclerosis in Chinese Han population. PLoS One. 2011;6(6):e20833.
Jarinova O, Stewart AF, Roberts R, Wells G, Lau P, Naing T, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29(10):1671–7.
Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429.
Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464(7287):409–12.
Gretarsdottir S, Baas AF, Thorleifsson G, Holm H, den Heijer M, de Vries JP, et al. Genome-wide association study identifies a sequence variant within the DAB2IP gene conferring susceptibility to abdominal aortic aneurysm. Nat Genet. 2010;42(8):692–7.
Shen YJ, Kong ZL, Wan FN, Wang HK, Bian XJ, Gan HL, et al. Downregulation of DAB2IP results in cell proliferation and invasion and contributes to unfavorable outcomes in bladder cancer. Cancer Sci. 2014;105(6):704–12.
Yu L, Shang ZF, Abdisalaam S, Lee KJ, Gupta A, Hsieh JT, et al. Tumor suppressor protein DAB2IP participates in chromosomal stability maintenance through activating spindle assembly checkpoint and stabilizing kinetochore-microtubule attachments. Nucleic Acids Res. 2016;44(18):8842–54.
Peng WH, Lu L, Zhang Q, Zhang RY, Wang LJ, Yan XX, et al. Chromosome 9p21 polymorphism is associated with myocardial infarction but not with clinical outcome in Han Chinese. Clin Chem Lab Med. 2009;47(8):917–22.
Li HL, Zhang HL, Jian WX, Li Q, Peng WH, Xu YW. Association of vaspin gene polymorphisms with coronary artery disease in Chinese population and function study. Clin Chim Acta. 2013;415:233–8.
Zhuang J, Peng W, Li H, Wang W, Wei Y, Li W, et al. Methylation of p15INK4b and expression of ANRIL on chromosome 9p21 are associated with coronary artery disease. PLoS One. 2012;7(10):e47193.
Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, et al. Heart disease and stroke Statistics-2017 update: a report from the American Heart Association. Circulation. 2017;135(10):e146–603.
McPherson R, Tybjaerg-Hansen A. Genetics of coronary artery disease. Circ Res. 2016;118(4):564–78.
Iannaccone M, Quadri G, Taha S, D'Ascenzo F, Montefusco A, Omede P, et al. Prevalence and predictors of culprit plaque rupture at OCT in patients with coronary artery disease: a meta-analysis. Eur Heart J Cardiovasc Imaging. 2016;17(10):1128–37.
Nasu K, Terashima M, Habara M, Ko E, Ito T, Yokota D, et al. Impact of cholesterol metabolism on coronary plaque vulnerability of target vessels: a combined analysis of virtual histology intravascular ultrasound and optical coherence tomography. JACC Cardiovasc Interv. 2013;6(7):746–55.
Koga S, Ikeda S, Yoshida T, Nakata T, Takeno M, Masuda N, et al. Elevated levels of systemic pentraxin 3 are associated with thin-cap fibroatheroma in coronary culprit lesions: assessment by optical coherence tomography and intravascular ultrasound. JACC Cardiovasc Interv. 2013;6(9):945–54.
Hiura Y, Fukushima Y, Yuno M, Sawamura H, Kokubo Y, Okamura T, et al. Validation of the association of genetic variants on chromosome 9p21 and 1q41 with myocardial infarction in a Japanese population. Circ J. 2008;72(8):1213–7.
Saleheen D, Alexander M, Rasheed A, Wormser D, Soranzo N, Hammond N, et al. Association of the 9p21.3 locus with risk of first-ever myocardial infarction in Pakistanis: case-control study in South Asia and updated meta-analysis of Europeans. Arterioscler Thromb Vasc Biol. 2010;30(7):1467–73.
Wang W, Peng W, Zhang X, Lu L, Zhang R, Zhang Q, et al. Chromosome 9p21.3 polymorphism in a Chinese Han population is associated with angiographic coronary plaque progression in non-diabetic but not in type 2 diabetic patients. Cardiovasc Diabetol. 2010;9:33.
Harrison SC, Cooper JA, Li K, Talmud PJ, Sofat R, Stephens JW, et al. Association of a sequence variant in DAB2IP with coronary heart disease. Eur Heart J. 2012;33(7):881–8.
Shahid SU, Shabana CJA, Beaney KE, Li K, Rehman A, et al. Genetic risk analysis of coronary artery disease in Pakistani subjects using a genetic risk score of 21 variants. Atherosclerosis. 2017;258:1–7.
Zhu XH, Wang JM, Yang SS, Wang FF, Hu JL, Xin SN, et al. Down-regulation of DAB2IP promotes colorectal cancer invasion and metastasis by translocating hnRNPK into nucleus to enhance the transcription of MMP2. Int J Cancer. 2017;141(1):172–83.
Acknowledgements
Not Applicable
Funding
None
Availability of data and materials
Raw data supporting the obtained results can be requested from the corresponding author.
Author information
Authors and Affiliations
Contributions
XL and YX conceived and designed the study. QL and HL participated in data acquisition. QL and JZ extracted DNA and performed genotype. QL and WP performed statistical analysis. All authors wrote and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of No. 113 Hospital of Chinese People’s Liberation Army. All patients gave written informed consent.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, Q., Peng, W., Li, H. et al. Association of the single nucleotide polymorphism in chromosome 9p21 and chromosome 9q33 with coronary artery disease in Chinese population. BMC Cardiovasc Disord 17, 255 (2017). https://doi.org/10.1186/s12872-017-0685-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12872-017-0685-0