Abstract
Background
Panicle architecture is one of the main important agronomical traits that determine branch number and grain number in rice. Although a large number of genes involved in panicle development have been identified in recent years, the complex processes of inflorescence patterning need to be further characterized in rice. Brassinosteroids (BRs) are a class of steroid phytohormones. A great understanding of how BRs contribute to plant height and leaf erectness have been reported, however, the molecular and genetic mechanisms of panicle architecture influenced by BRs remain unclear.
Results
Here, we identified PMM1, encoding a cytochrome P450 protein involved in BRs biosynthesis, and characterized its role in panicle architecture in rice. Three alleles of pmm1 were identified from our T-DNA insertional mutant library. Map-based cloning revealed that a large fragment deletion from the 2nd to 9th exons of PMM1 was responsible for the clustered primary branch morphology in pmm1–1. PMM1 is a new allele of DWARF11 (D11) PMM1 transcripts are preferentially expressed in young panicles, particularly expressed in the primordia of branches and spikelets during inflorescence development. Furthermore, overexpression of OsDWARF4 (D4), another gene encoding cytochrome P450, completely rescued the abnormal panicle phenotype of pmm1–1. Overall, it can be concluded that PMM1 is an important gene involved in BRs biosynthesis and affecting the differentiation of spikelet primordia and patterns of panicle branches in rice.
Conclusions
PMM1 is a new allele of D11, which encodes a cytochrome P450 protein involved in BRs biosynthesis pathway. Overexpression of D4 could successfully rescue the abnormal panicle architecture of pmm1 plants, indicating that PMM1/D11 and D4 function redundantly in BRs biosynthesis. Thus, our results demonstrated that PMM1 determines the inflorescence architecture by controlling brassinosteroid biosynthesis in rice.
Similar content being viewed by others
Background
Inflorescence architecture and spikelet formation are unique features of grasses such as rice, maize and wheat. In rice, inflorescence (also called panicle) architecture can be categorized into nine successive stages according to the morphological dynamic changes [1]. During the transition from vegetative to reproductive phase, inflorescence meristem (IM) firstly initiates from shoot apex meristem (SAM), and subsequently produces primary, secondary, and sometimes a higher order of panicle branch meristem (BM). On lateral panicle branches, spikelet meristem (SM) initiates and subsequently bears florets and finally develops into grain [1,2,3]. As a result, the length of the main axis as well as the number/length of primary and secondary inflorescence branches are two major determinants for the number of spikelets per panicle and yield of rice [4].
In recent years, multiple genes have been identified to determine inflorescence development processes and finally form the morphology of panicle. SMALL PANICLE (SPA), REDUCED CULM NUMBER1 (RCN1), LAX PANICLE1 (LAX1), and LAX2 are involved in the initiation of BM and SM [5,6,7,8,9]. ABERRANT PANICLE ORGANIZATION1 (APO1), APO2/RFL and TAWAW1 (TWA1) are responsible for the identity of BM by preventing precocious conversion of BM to SM [10,11,12,13,14,15,16]. FRIZZLE PANICLE (FZP) is required for floral organ initiation and identity through preventing formation of axillary meristems of rice spikelets [17,18,19]. TILLERS ABSENT1 (TAB1) has been shown to be involved in the activity of axillary meristems in rice [20, 21]. Some other genes, such as GRAIN NUMBER1 (Gn1a), DENSE AND ERECT PANICLE1 (DEP1), DEP2 and DEP3, showed effects on the number of branches or spikelets in rice [22,23,24,25,26,27]. SHORT PANICLE1 (SP1), ABERRANT SPIKELET AND PANICLE1 (ASP1), PANICLE APICAL ABORTION1 (PAAB1), TUTOU1 (TUT1) and SQUAMOSA PROMOTERBINDING PROTEIN-LIKE 6 (SPL6) have been identified to be involved in the elongation of inflorescence branches and degeneration of panicles in rice [28,29,30,31,32]. Besides, a few genes involved in panicle density have also been reported. OsLG1/SPR3 encodes a SBP-domain transcription factor and regulates a closed panicle trait, a selected trait during rice domestication [33,34,35]. In addition, CL and CL-DZ have been described as genes that lead to typical clustered spikelets and are necessary for the formation of bract primordia in the primary and secondary branch meristems [36, 37]. Our previous investigation has identified a panicle morphology mutant 1 (pmm1), which also causes a clustered primary branches in panicle [38]. CLUSTERED PRIMARY BRANCH1 (CPB1) was identified to influence the development of panicle architecture, leaf angle and seed size [39]. Despite of these advances, the molecular and genetic mechanisms underlying the differentiation of spikelets or branch meristems are still poorly understood.
Brassinosteroids (BRs) are a class of steroid phytohormones, which influence both plant height and leaf erectness in rice [40]. So far, several genes playing crucial roles in BRs biosynthesis or signaling pathways have been identified via mutants either BR-deficient or BR-insensitive. The BR-insensitive mutants showed no significant response to the exogenous application of BRs due to the loss-of-function of the genes involved in the signal transduction pathway for BRs, such as OsBRI1/DWARF61, DWARF1, OsBZR1, 14–3-3, OsBAK1, OsBRL, SG1, XIAO, DWARF62, and OsGRAS19 [41,42,43,44,45,46,47,48,49,50]. BR-deficient mutant, such as dwarf2 (d2), dwarf4 (d4), dwarf11 (d11), brd1 and brd2, showed defects in BR biosynthesis, but the defects could be rescued by exogenous application of BRs [51,52,53,54,55,56,57]. Both DWARF4 (D4) and DWARF11 (D11) encode a cytochrome P450, which involved in BRs biosynthesis and have been characterized to control the plant architecture [39, 54]. D4 encodes CYP90B and catalyzes C-22 hydroxylation, which is a rate-limiting step of BRs biosynthesis [58, 59]. Mutation of D4 caused defects in rice plant morphology, such as a slight dwarfed stature and more erect leaves, indicating that D4 may mainly function during vegetative organs in rice [54]. D11, which encodes CYP724B1, is essential to maintain the levels of bioactive BRs synthesis [55]. d11 mutant showed the pleiotropic defective morphologies including erect leaves, shortened internodes and small seeds [55]. However, the expression patterns of D11 and D4 and their roles in panicle architecture need to further elucidate.
In this study, we identified two additional alleles of pmm1, which exhibited a clustered primary branches phenotype during panicle development. Our genetic mapping results revealed that PMM1 is a new allele of D11, which encodes a cytochrome P450 protein involved in BRs biosynthesis pathway. Transgenic analysis indicated that PMM1/D11 and D4 function redundantly in BRs biosynthesis, indicating that BRs biosynthesis is required for the panicle architecture in rice.
Results
Identification of pmm1 mutants
Previously, we have identified a panicle morphology mutant 1 (pmm1) [38], which showed a slight phenotype in vegetative growth (Fig. 1a) but a strong morphological defects in inflorescence architecture (Fig. 1b). We designated this panicle morphology mutant as pmm1–1. The panicle morphology of pmm1–1 was dramatically altered compared with that of wild type (WT), such as clustered primary branching, opposite grains, and small grains (Fig. 1b). In order to collect more panicle morphology mutants, we screened our T-DNA insertional mutant library [60] and identified two additional lines, which showed obvious clustered primary branches similar to pmm1–1 (Fig. 1a, b). Genetic analyses showed that about one- quarter of their progenies of the heterozygous were defect panicle morphology and others showed normal panicle, indicating that the clustered primary branching of each mutant was controlled by a single recessive allele, respectively. We did not detect any phenotypic segregation when crossing each mutant with pmm1–1 (Fig. 1c), suggesting that the mutation loci in these two independent lines were allelic to that in pmm1–1. Thus, these two lines were designated as pmm1–2 and pmm1–3, respectively. We chose pmm1–1 for further examination.
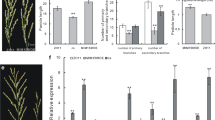
Phenotypic characterization of pmm1 mutants. a Plant morphology between WT (ZH11) and three alleles of pmm1 mutants. b Panicle architecture between WT and three alleles of pmm1 mutants. c Plant morphology of F1 plants of pmm1–1/pmm1–2 and pmm1–1/pmm1–3. d Spikelet morphology between WT pmm1–1 mutants. e-h Comparison of the primary branch number (PBs) (e), secondary branch number (SBs) (f), grain number per panicle (g) and 1000-grains weight (h) between WT and pmm1–1. Data are shown as means ± SE (n = 10). Significant at **P < 0.01 and *P < 0.05
Characterization of pmm1
Next, we characterized the morphological defects of pmm1 compared with WT. In the paddy field, all the pmm1 plants (pmm1–1, pmm1–2 and pmm1–3) appeared slightly dwarf and produced erected leaves (Fig. 1a). After heading, pmm1 plants showed obvious multiple morphological defects in panicle, such as clustered panicle, shortened axis, clustered primary branches and opposite spikelets (Fig. 1b, d). We further compared the yield traits between pmm1–1 and the WT plants under normal growth conditions. The primary branches number per panicle was increased in pmm1–1 (Fig. 1e), while as for the other traits, no obvious differences concerning secondary branches number per panicle and grain number per panicle in pmm1–1 compared with WT (Fig. 1f, g). A 30% decrease in 1000-grain weight was decreased in pmm1–1 (Fig. 1h).
To better characterize the panicle developmental defects of pmm1–1, we compared the panicle development between WT and pmm1–1 using scanning electron microscopy (SEM). No remarkable morphological differences in apices were observed between pmm1–1 and WT (Fig. 2a, e). At the reproductive stage, the primary branches were generated normally in WT (Fig. 2b). However, the primary branch primordia in pmm1–1 seemed to generate in a whorl pattern (Fig. 2f). At the secondary branch primordial formation stage, no obvious differences were observed between pmm1–1 and WT (Fig. 2c, g). At the spikelet formation stage, the spikelet primordia were normally developed in WT (Fig. 2d) and pmm1–1 (Fig. 2h). Compared to WT (Fig. 2i, j), histological analysis revealed that such malformation would due to the clustered primary branches pattern developed in pmm1–1 (Fig. 2k, l). These observations suggested that the generation pattern of primary branches were altered in pmm1–1 compared with WT.
Microscopic observation of the developing panicles in WT and pmm1–1. Scanning electron microscopy (SEM) analysis of young panicle development between WT (a-d) and pmm1–1 (e-h) at the vegetative stage (a, e), the primary branch primordial formation stage (b, f), the second primordial formation stage (c, g), the spikelet primordial formation stage (d, h). Longitudinal section analysis of young panicle development in WT (i, j) and pmm1–1 (k, l) at the primary branch primordial formation stage (i, k) and the second primordial formation stage (j, l). Bars = 100 μm
Identification of PMM1
Previously, we have already generated a F2 mapping population by crossing pmm1–1 with an indica variety ZS97 [38] and delimited PMM1 to a 147 kb region between the two markers RM3866 and X4 (Fig. 3a). Since no more polymorphic markers could be developed to further narrow down the candidate locus (Additional file 1: Table S1), we sequenced and analyzed all the 21 predicted genes in this region (Additional file 2: Table S2). Finally, we identified a 4124 bp deletion in this region (Fig. 3b), which resulted in the deletion of a large fragment from 2nd to 9th exons of LOC_Os04g39430 (D11) (Fig. 3c). Thus, pmm1–1 mutant might be a knock-out mutant of D11. Further analysis showed that the alteration of panicle architecture in pmm1–1 mutant were not associated with the T-DNA insertion (Fig. 3d), suggesting that the large fragment deletion might have occurred due to tissue culture process. qRT-PCR results revealed that the transcripts of D11 were significantly blocked in the young panicles of pmm1–1 compared with WT (Fig. 3e). Therefore, D11 is likely to be a candidate for PMM1.
Map-based cloning of PMM1. a Linkage map of PMM1 on chromosome 4. The number of recombinants between the molecular marker and PMM1 is indicated. b Location of PMM1. c Gene structure of PMM1. The black boxes represent the exons and lines between the boxes represent introns. The deletion fragment in pmm1–1 is shown. d Genotyping of ZH11 and pmm1–1 by PCR analysis. e Transcript levels of PMM1 in WT and pmm1–1 determined by qRT-PCR. Rice UBIQIUTIN5 was used as an internal control. Data are presented as means ± SE (n = 3). Significant at **P < 0.01
Genetic complementation of pmm1
To confirm that D11 is responsible for the phenotype of pmm1–1, we generated a vector containing the D11-coding region under the control of its native promoter and transformed it into pmm1–1 mutant by Agrobacterium tumefaciens-mediated transformation [60]. Forty-seven independent transgenic plants were generated, and the clustered primary branches of pmm1–1 were completely restored to normal inflorescence architecture in all the transgenic positive plants (Fig. 4a, b). Besides, the grain length and grain width were increased compared to the negative plants (Fig. 4c, d), the grain size also enlarged as much as that in WT. Collectively, these results suggest that the mutation in D11 is responsible for abnormal panicle morphology of pmm1–1.
Genetic complementation of pmm1–1. a Comparison of plant phenotypes between negative and positive transgenic plants. b Panicle architecture of transgenic plants. c Grain width of transgenic plants. d Grain length of transgenic plants. C-N, negative transgenic plants; C-P, positive transgenic plants. Bars = 0.5 cm
PMM1 encodes a cytochrome P450 involved in BRs biosynthesis
D11 encodes a cytochrome P450 superfamily protein CYP724B1, facilitating the supply of 6-DeoxoTY and TY during BRs biosynthesis [55]. As PMM1 was identified as a new allele of D11, we speculated that pmm1 would be deficient in BRs biosynthesis and sensitive to BRs. To confirm that, we carried out morphological examination of pmm1–1 in the complete darkness. The emergence rate of mesocotyls and the length of coleoptile were investigated in pmm1–1 and WT plants. In the dark, the WT plants showed an obvious skotomorphogenic phenotype including elongated mesocotyls and coleoptile (Fig. 5a). However, the mesocotyls were not elongated and the length of coleoptile was short in pmm1–1 (Fig. 5b). Then, we treated pmm1–1 plants with the most bioactive BRs compound BL in darkness. The emergence rate of mesocotyls in pmm1–1 was increased with improving the concentration of BL (Fig. 5c). The length of coleoptile in pmm1–1 was almost the same as that in WT under the concentration of 10− 6 M (Fig. 5d). Our result confirmed that pmm1–1 is a BR-sensitive mutant and PMM1 might be involved in BRs biosynthesis.
Response of seedlings to BL. a Skotomorphogenesis of WT plants treated with BLs. b Skotomorphogenesis of pmm1–1 plants treated with BLs. c Rate of mesocotyls emergence between WT and pmm1–1 plants. d Length of coleoptiles between WT and pmm1–1 plants. The positions of mesocotyls in these plants are indicated by red stars. The lengths of coleoptile in these plants are indicated by white arrows. Data are presented as means ± SE (n = 10). Significant at **P < 0.01
PMM1 transcripts are abundant in young panicle
To examine the PMM1 expression pattern, we firstly conducted RT-PCR assays on the RNA samples from root, stem, leaf blade, leaf sheath and developing panicles. PMM1 transcripts were detected at a very low level in the examined vegetative organs, but highly accumulated in the developing panicles (Fig. 6a). Subsequently, in situ hybridization was carried out to determine the precise expression pattern in the young panicles. The RNA in situ hybridization signals was obviously detected in the branch primodia and the spikelet primodia in the developing young panicle (Fig. 6b). However, the PMM1 signals were absent in pmm1–1 (Fig. 6c). Using the sense-strand probes, no signal was detected either in pmm1–1 (Fig. 6d) or in WT (Fig. 6e). Because PMM1 preferentially expressed in young panicle and its mutant showed panicle defects, we deduced that PMM1 would be required for the panicle architecture in rice.
Expression pattern of PMM1. a Expression profiles of PMM1 in the root, stem, leaf, leaf sheath and developing panicles at the 1, 2, 3.5, 4.5, 6.5, 8, 10 and 16.5-cm stages before heading. Rice UBIQIUTIN5 was used as an internal control. Data are presented as means ± SE (n = 3). b-e In situ hybridization assay of PMM1 in young panicles of WT (b) and pmm1–1 (c). Negative controls with sense probe in in young panicles of WT (d) and pmm1–1 (e). Bars = 100 μm
Overexpression of OsDWARF4 could rescue the abnormal panicle architecture of pmm1
OsDWARF4 (D4) has been shown to function redundantly with D11 in BRs biosynthesis [54]. In order to examine the roles of D4 in the panicle architecture in rice, we firstly conducted the sequences comparison between D4 and D11. Sequence alignment showed that the amino acid sequence of D11 shared more than 40% identity with D4 (Additional file 3: Figure S1a). Then, we performed qRT-PCR analysis to examine the expression patterns of D4 in different tissues of both pmm1–1 and WT. The results showed that D4 was highly expressed in the root, culm, leaf and leaf sheath but lowly expressed in the developing panicle (Additional file 3: Figure S1b). The transcript levels of D4 were increased in pmm1–1 plants, suggesting that D4 expression might be feedback-regulated when loss of function of PMM1/D11.
To confirm the functional redundancy between D4 and PMM1/D11, we generated 30 plants overexpressing D4 in pmm1–1 and in Zhonghua 11 (control), respectively. qRT-PCR analysis confirmed the positive transgenic lines (D4-OE, D4-OE/ pmm1–1) had overexpression of D4 in young panicles compared with the negative transgenic lines (Fig. 7a). D4-overexpressing plants (e.g. D4-OE #8) exhibited larger leaf angles, a typical phenotypic characteristic of excess BRs (Fig. 7b). In the plants of D4-OE/pmm1–1 (#13), plant height and the abnormal panicle morphology could be completely recovered by overexpression of D4 expression in pmm1–1 (Fig. 7b, c). We have also generated 48 plants overexpressing PMM1/D11. qRT-PCR analysis confirmed the overexpression of PMM1 in the independent positive transgenic lines compared with the negative transgenic lines (Fig. 7d). The plants overexpressing PMM1 (#12) displayed large leaf angles (Fig. 7e, g) and increase in plant height (Fig. 7e, h), but a decrease in panicle length (Fig. 7f, i). We have detected a slight increase grain number per panicle and 1000-grain weight in PMM1 overexpression lines compared with control (Fig. 7j). Taken together, our result suggested that D4 and PMM1/D11 function redundantly in biosynthesis of BRs, which is crucial for the normal panicle architecture development.
Morphological characterization of D4-overexpressing and PMM1-overexpressing plants. a Transcript levels of D4 in positive (P) and negative (N) transgenic plants in WT and pmm1–1 backgrounds, respectively. Rice UBIQIUTIN5 was used as an internal control. Data are presented as means ± SE (n = 3). b Plant morphology of D4-overexpressing lines. c Panicle morphology of D4-overexpressing lines. d Transcript levels of PMM1 in positive (P) and negative (N) transgenic plants. Rice UBIQIUTIN5 was used as an internal control. Data are presented as means ± SE (n = 3). e Plant morphology of PMM1-overexpressing lines. f Panicle morphology of PMM1-overexpressing lines. g Flag leaf angles of D4-overexpressing lines. h Plant height of D4-overexpressing lines. i Panicle length of D4-overexpressing lines. j 1000-grains weight of D4-overexpressing lines. Data are presented as means ± SE (n = 3). Significant at **P < 0.01
PMM1 may participate in multiple biological processes
Previous works have elucidated that PMM1/D11 encodes a cytochrome P450 involved in BRs biosynthesis process [54, 55]. In order to investigate the possible roles of PMM1 in BR biosynthesis, we examined the expression levels of one BR biosynthesis gene (D4) and seven BR signal transduction genes (OsMDP1, BU1, ILI1, RAVL1, OsBRI1, OsLIC and SG1) in the young panicles of pmm1–1 by qRT-PCR. Compared to that of WT, the expression levels of BR-signaling genes did not altered significantly in pmm1–1 except for MDP1, which was significantly reduced in pmm1–1 (Fig. 8a). As reported in previous literature, the expression level of the BRs biosynthesis gene D4 was significantly increased in d11/pmm1 (Fig. 8a), indicating that a feedback regulation of BRs biosynthesis genes may be trigged in d11/pmm1 [54]. In the D4-OE/WT plants, the expression levels of some BR signal transduction genes (such as OsBRI1, OsMDP1, and BU1) showed dramatically increased compared to the WT (Fig. 8b). These results further indicated that D11 and D4 might be responsible for BR biosynthesis in different organs. Although overexpression of D4 in the young panicles could rescue the clustered panicles in pmm1, the dosage effect of BRs might cause the different expression patterns of BR signal transduction genes in D4-OE/pmm1 plants (Fig. 8b).
Expression pattern of corresponding genes in the young panicles of WT, pmm1–1 and D4-overexpressing plants. a Expression pattern of BR-related genes in WT and pmm1–1 plants. b Expression pattern of BR-related genes in D4-overexpressing plants. c Expression levels of several genes related to panicle development in WT and pmm1–1 plants. d Expression levels of several genes related to cell cycling. Rice UBIQIUTIN5 was used as an internal control. Data are presented as means ± SE (n = 3). Significant at **P < 0.01 and *P < 0.05
LAX1, LAX2 and FZP are classic genes, which determine the inflorescence architecture by controlling rachis-branch meristem development [6, 9, 17]. We have examined whether the panicle architecture regulatory pathway is associated with the PMM1/D11 using qRT-PCR analysis in young panicle. The expression levels of FZP and LAX2 were not significantly changed in pmm1–1, suggesting that PMM1 does not affect the expression of FZP and LAX2 (Fig. 8c). In contrast, the expression of LAX1 was significantly up-regulated in pmm1–1, indicating that BRs might be involved in LAX1 regulatory pathways which influence panicle formation in rice.
Previous studies have suggested that BRs can influence seed sizes by affecting cell cycling genes in rice [49]. In this study, mutation in PMM1 also caused small and round grains compared with WT (Fig. 1b). Then, we analyzed the expression levels of six cell cycle-related genes (MCM2, MCM3, CYCB2, CYCB2.1, CYClaZm, CYCA2.3) in young panicles of pmm1–1 and WT. The expression of both MCM2 and MCM3 significantly increased in pmm1–1 compared with that in WT, but that of the other cell cycle-related genes (CYCB2, CYCB2.1, CYClaZm, CYCA2.3) were not significantly changed (Fig. 8d). These results suggested that PMM1, a BRs biosynthesis gene, might contribute a possible connection between BRs and cell cycle regulation in controlling grain size in rice.
Discussion
Mutation in PMM1 resulted in clustered panicles
Panicle morphology is a critical determinant of grain yield in rice and other grain crops. So far, diverse inflorescence architectures have been identified in rice, such as short panicle [28], long panicle [61], large panicle [62], lax panicle [6, 7, 9], frizzy panicle [17,18,19], dense panicle [23,24,25], abortive panicle [29,30,31], spreading panicle [33,34,35], and clustered panicle [39], which would be potential genetic resources for breeding of an ideal rice panicle morphology. Identification of superior alleles for panicle architecture and unraveling their molecular basis would shed light on future breeding program.
In this study, we identified 3 independent lines with mutations in pmm1 exhibiting whorled primary branches and clustered spikelets (Fig. 1c, d). SEM results showed that the clustered branch precordia occurred during the young panicle development in pmm1–1 (Fig. 1). Therefore, we hypothesized that PMM1 plays an important role in determination of branch and spikelet primordium formation in inflorescences. In addition, expression data showed that PMM1 was predominantly expressed in developing young inflorescence, particularly in branches and spikelet primordia (Fig. 6b). We speculate that PMM1 largely affects the panicle growth and development in the early stages of axillary meristem initiation. Considering pmm1–2 and pmm1–3 are allelic to pmm1–1, we have not further characterized their panicle morphology in detail. Actually, by screening our T-DNA insertion mutant library, we have collected more than 50 independent mutant lines that showed obvious altered panicle architecture. However, only these 3 independent lines (pmm1–1, pmm1–2, and pmm1–3) caused an obvious clustered primary branches in panicle. Previous literature have reported that CPB1 gene, an allele of DWRAF11, also controls panicle architecture and seed size in rice [39]. Mutation in DWRAF11 resulted in remarkable clustered branches in panicles, indicating that BR biosynthesis in panicle is essential for normal inflorescence architecture in rice.
PMM1 encodes a cytochrome P450 related to BR biosynthesis
Physiological studies have demonstrated that most BR-biosynthesis genes are involved in diverse processes during plant development, such as stem elongation, leaf angle, tiller number, plant height, male fertility, senescence and biotic/abiotic stresses [63, 64]. In this study, loss function of PMM1 also resulted in a typical phenotype of BR biosynthesis deficiency, such as compact plant type with reduced leaf angle, small and round grains (Fig. 1b, d). Actually, we also observed the reduced leaf angles and shortened internodes in pmm1–1 compared to WT (Additional file 4: Figure S2 a-g). Additionally, the expression levels of genes associated with lamina joint inclination and BR-related genes associated with internode elongation have been changed in pmm1–1 plants (Additional file 4: Figure S2 h, i). The pmm1–1 plants showed a highly sensitivity to BLs treatment (Fig. 5). These results strongly suggest that PMM1 encodes a cytochrome P450 and participates in BR biosynthesis pathway [55].
Besides the typical characteristic phenotypes of BR deficiency, the pmm1–1 mutant, with the deletion of a large fragment from 2 to 9 exons, displayed a novel phenotype of whorled branches and clustered spikelets, with 2–3 abnormal spikelets clustering on each panicle branch (Fig. 1d). PMM1 is a novel allele of DWARF11. Previous studies found that some allelic D11 mutations were generated because of single nucleotide deletion (d11–1), insertion (d11–2) or substitution (d11–3, d11–4 and cpb1) [39, 55]. Nevertheless, none of these mutants exhibited whorled primary branches and clustered spikelets, except for that cpb1 showed clustered primary branches but no obvious clustered spikelets [39]. These results suggest that Oryza sativa cytochrome P450 family member CYP724B1 affects plant architecture, especially inflorescence architecture, might be in a transcript dosage-dependent manner, or depending on rice varieties.
Many other kinds of BR-related genes have also been demonstrated to affect rice plant architecture, such as BRD1, BRD2 and XIAO [49, 51, 53, 56]. Among them, XIAO was reported to be involved in the control of organ size by cell cycling. We have observed the small and round grains in three pmm1 mutants (Fig. 1b). Expression analysis showed that there were no significant changes in gene expression between WT and pmm1–1, except for MCM2 and MCM3 (Fig. 8d). These results suggest that PMM1/D11 controls the seed sizes possibly through cell cycling as well.
The relationship between panicle architecture and genes involved in BRs signaling and biosynthesis
It is well known that there is an interplay between the signaling and biosynthesis of BRs in rice [49]. However, some of these tested BR-related genes, including RAVL1, OsBRI1, OsLIC and SG1, did not show conspicuous alterations in pmm1–1 except for MDP1 and DWARF4 (Fig. 8a). A previous study suggested that OsMDP1 function in BR signal transduction and act as a negative regulator of floral organ development and floral identity in rice [65]. To investigate whether panicle architecture in rice requires OsMDP1, a closer examination of panicle morphology with osmdp1 need to be carried out.
As previously reported, D11 and D4 may function redundantly in C-22 hydroxylation for BR biosynthesis [54]. We have verified that overexpression of D4 under the background of pmm1–1 mutant could completely rescue the abnormal inflorescence in pmm1–1 (Fig. 7c). These results indicate that enhancing the expression level of D4 could complement the BR deficit in pmm1–1. However, in plants with gain or loss function of D4, no significant variations could be observed in branch and spikelet morphology (Fig. 7c), indicating that D4 is not crucial to the establishment of panicle morphology in rice. These results suggest that PMM1 acts on the establishment of rice panicle architecture by regulating BR accumulation, while D4 might contribute to vegetative development preferentially.
Conclusion
We have identified a new allele of D11, PMM1, encoding a cytochrome P450 protein involved in BR biosynthesis pathway. A large fragment deletion in pmm1–1 caused an obvious change in rice panicle morphology. Our research revealed that BR biosynthesis is required for the primordial initiation of branches and spikelets during the young panicle development. Thus, PMM1 determines the inflorescence architecture of rice by controlling brassinosteroid biosynthesis.
Methods
Plant materials and growth conditions
All rice plants used in this study were japonica (O. sativa ssp. geng) variety Zhonghua 11 (ZH11). Three pmm1 mutants were obtained by screening the enhancer trap mutant library [60]. Rice plants were cultivated in the experimental field of Huazhong Agriculture University in the normal growing season in Wuhan, China (latitude 30.5°N, 15 m above sea level; average daily temperature approximately 28 °C).
Plasmid construction and rice transformation
To prepare the complementation vector, we digested ZH11 BAC clone OSJNBa0020J04 (kindly provided by Luo Meizhong) with EcoRI and XbaI, and a 6.2-kb genomic DNA fragment containing the entire PMM1 coding region and the 1600-bp upstream and 800-bp downstream sequences was inserted into pCAMBIA2301. The empty vector pCAMBIA2301 (http://www.cambia.org/daisy/cambia/585) was used as a negative control. For overexpression of PMM1/D11 or OsDWARF4, full-length cDNA of PMM1/D11 or DWARF4 was cloned into the pU1301 vector, which was then electroporated into the Agrobacterium tumefaciens strain EHA105, and finally transformed into rice callus to generate transgenic plants.
Scanning electron microscopy
For scanning electron microscopy, young panicles at different developmental stages from WT and pmm1–1 mutants were dissected, subsequently fixed with a blade, and immediately placed in 70% ethanol, 5% acetic acid, and 3.7% formaldehyde for 24 h at 4 °C overnight. Tissues were dehydrated through an ethanol series of 25 to 100% and dried. Following ethanol dehydration, the samples were critical point dried, sputter coated with gold in an E-100 ion sputter, and then observed under a scanning electron microscope (S570, Hitachi, Tokyo, Japan).
RNA extraction and gene expression analysis
Total RNA was extracted from frozen samples using TRIZOL reagent (Invitrogen). After RNase-free DNase I treatment, the first-strand cDNA was synthesized from 4 μg of total RNA with oligo (dT)15 as the primer, using a reverse transcription kit (Invitrogen). qRT-PCR was carried out using ABI7500 real-time PCR system with the SYBR Premix Ex Taq (TaKaRa) following the manufacturer’s instructions. The rice UBIQUITIN5 gene was used as an internal control. Gene expression level was determined from three independent replicates, each consisting of three plants, and three technical replicates per tissue sample were analyzed. The primers for real-time PCR are listed in Additional file 5: Table S3.
BR test
The WT and pmm1–1 mutant were used for BR induction experiments. Rice seeds were sterilized with 0.1% HgCl2 and then grown on 1% agar medium containing half-strength MS medium in complete darkness at 28 °C for 10 days. For BL (Wuhan DingGuo Biotech. Co. Ltd) induction of shoot elongation, rice seeds were grown on 1% agar medium containing half-strength MS medium and various concentrations (10− 7 M and 10− 6 M) of BL and incubated at 28 °C under continuous light. After 2 weeks, the length of coleoptile and the appearance of hypocotyl were measured. A total of 45 plants were used for each treatment.
In situ hybridization
RNA in situ hybridization was performed as described previously [66]. The probe was PCR-amplified from ZH11 using primers in situ-PMM1 F/R (Additional file 5: Table S3). The sense and antisense probes were then transcribed in vitro from the T7 or SP6 promoter with polymerase using a digoxigenin RNA labeling kit (Roche).
Abbreviations
- BM:
-
Branch meristem; SM: Spikelet meristem
- BRs:
-
Brassinosteroids
- D11 :
-
DWARF 11
- D4 :
-
DWARF 4
- PMM1:
-
Panicle Morphology Mutant 1
- SAM:
-
Shoot apex meristem
References
Ikeda K, Sunohara H, Nagato Y. Developmental course of inflorescence and spikelet in Rice. Jpn J Breeding. 2004;54(2):147–56.
Xing Y, Zhang Q. Genetic and molecular bases of rice yield. Annu Rev Plant Biol. 2010;61(1):421–42.
Zhang D, Yuan Z. Molecular control of grass inflorescence development. Annu Rev Plant Biol. 2014;65(1):553–78.
Wang Y, Li J. Branching in rice. Curr Opin Plant Biol. 2011;14(1):94–9.
Nakagawa M, Shimamoto K, Kyozuka J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002;29(6):743–50.
Komatsu M, Maekawa M, Shimamoto K, Kyozuka J. The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis–branch and spikelet development. Dev Biol. 2001;231(2):364–73.
Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J. LAX and SPA: major regulators of shoot branching in rice. Proc Natl Acad Sci U S A. 2003b;100(20):11765–70.
Oikawa T, Kyozuka J. Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell. 2009;21(4):1095–108.
Tabuchi H, Zhang Y, Hattori S, Omae M, Shimizu-Sato S, Oikawa T, Qian Q, Nishimura M, Kitano H, Xie H, et al. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell. 2011;23(9):3276–87.
Kyozuka J, Konishi S, Nemoto K, Izawa T, Shimamoto K. Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. Proc Natl Acad Sci U S A. 1998;95(5):1979–82.
Ikeda K, Ito M, Nagasawa N, Kyozuka J, Nagato Y. Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J. 2007;51(6):1030–40.
Rao NN, Prasad K, Kumar PR, Vijayraghavan U. Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc Natl Acad Sci U S A. 2008;105(9):3646.
Terao T, Nagata K, Morino K, Hirose T. A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice. Theor Appl Genet. 2009;120(5):875–93.
Ookawa T, Hobo T, Yano M, Murata K, Ando T, Miura H, Asano K, Ochiai Y, Ikeda M, Nishitani R, et al. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat Commun. 2010;1:132.
Ikeda-Kawakatsu K, Maekawa M, Izawa T, Itoh J, Nagato Y. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 2012;69(1):168–80.
Yoshida A, Sasao M, Yasuno N, Takagi K, Daimon Y, Chen R, Yamazaki R, Tokunaga H, Kitaguchi Y, Sato Y, et al. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc Natl Acad Sci U S A. 2013;110(2):767–72.
Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development. 2003;130(16):3841–50.
Bai X, Huang Y, Mao D, Wen M, Zhang L, Xing Y. Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Sci Rep. 2016;6:19022.
Bai X, Huang Y, Hu Y, Liu H, Zhang B, Smaczniak C, Hu G, Han Z, Xing Y. Duplication of an upstream silencer of FZP increases grain yield in rice. Nat Plants. 2017;3(11):885–93.
Lu Z, Shao G, Xiong J, Jiao Y, Wang J, Liu G, Meng X, Liang Y, Xiong G, Wang Y, et al. MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J Genet Genomics. 2015;42(2):71–8.
Tanaka W, Ohmori Y, Ushijima T, Matsusaka H, Matsushita T, Kumamaru T, Kawano S, Hirano HY. Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1. Plant Cell. 2015;27(4):1173–84.
Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles E, Qian Q, Kitano H, Matsuoka M. Cytokinin oxidase regulates rice grain production. Science. 2005;309(5735):741–5.
Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet. 2009;41(4):494–7.
Li F, Liu W, Tang J, Chen J, Tong H, Hu B, Li C, Fang J, Chen M, Chu C. Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res. 2010;20(7):838–49.
Qiao Y, Piao R, Shi J, Lee SI, Jiang W, Kim BK, Lee J, Han L, Ma W, Koh HJ. Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.). Theor Appl Genet. 2011;122(7):1439–49.
Sun H, Qian Q, Wu K, Luo J, Wang S, Zhang C, Ma Y, Liu Q, Huang X, Yuan Q, et al. Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat Genet. 2014;46(6):652–6.
Liu Q, Han R, Wu K, Zhang J, Ye Y, Wang S, Chen J, Pan Y, Li Q, Xu X, et al. G-protein betagamma subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat Commun. 2018;9(1):852.
Li S, Qian Q, Fu Z, Zeng D, Meng X, Kyozuka J, Maekawa M, Zhu X, Zhang J, Li J, et al. Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 2009;58(4):592–605.
Yoshida A, Ohmori Y, Kitano H, Taguchi-Shiobara F, Hirano HY. Aberrant spikelet and panicle1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 2012;70(2):327–39.
Bai J, Zhu X, Wang Q, Zhang J, Chen H, Dong G, Zhu L, Zheng H, Xie Q, Nian J, et al. Rice TUTOU1 encodes a suppressor of cAMP receptor-like protein that is important for actin organization and panicle development. Plant Physiol. 2015;169(2):1179–91.
Heng Y, Wu C, Long Y, Luo S, Ma J, Chen J, Liu J, Zhang H, Ren Y, Wang M, et al. OsALMT7 maintains panicle size and grain yield in rice by mediating malate transport. Plant Cell. 2018;30(4):889–906.
Wang QL, Sun AZ, Chen ST, Chen LS, Guo FQ. SPL6 represses signalling outputs of ER stress in control of panicle cell death in rice. Nat Plants. 2018;4:280–8.
Luo JJ, Hao W, Jin J, Gao JP, Lin HX. Fine mapping of Spr3, a locus for spreading panicle from African cultivated rice (Oryza glaberrima steud.). Mol Plant. 2008;1(5):830–8.
Zhu Z, Tan L, Fu Y, Liu F, Cai H, Xie D, Wu F, Wu J, Matsumoto T, Sun C. Genetic control of inflorescence architecture during rice domestication. Nat Commun. 2013;4(3):2200.
Ishii T, Numaguchi K, Miura K, Yoshida K, Thanh PT, Htun TM, Yamasaki M, Komeda N, Matsumoto T, Terauchi R, et al. OsLG1 regulates a closed panicle trait in domesticated rice. Nat Genet. 2013;45(4):462–5.
Murai M, Lizawa M. Effects of major genes controlling morphology of panicle in rice. Breed Sci. 2010;44(3):247–55.
Tian C, Zhang T, Jiang KF, Yang L, Yang QH, Wan XQ. Genetic analysis and preliminary gene mapping of rice clustered spikelet mutant. Mol Plant Breed. 2010;8(1):29–34.
Li X, He Z, Yu S, Chen G, Wu C. Genetic analyses and preliminary mapping of a panicle morphogenesis mutant (pmm1) in rice (in Chinese). J HZAU. 2013;32:10–5.
Wu Y, Fu Y, Zhao S, Gu P, Zhu Z, Sun C, Tan L. CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol J. 2016;14(1):377–86.
Tong H, Chu C. Brassinosteroid signaling and application in rice. J Genet Genomics. 2012;39(1):3–9.
Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A. Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc Natl Acad Sci U S A. 1999;96(18):10284–9.
Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000;12(9):1591–605.
Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci U S A. 2007;104(34):13839–44.
Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH, Li J, Chong K. Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J. 2009;7(8):791–806.
Nakamura A, Fujioka S, Sunohara H, Kamiya N, Hong Z, Inukai Y, Miura K, Takatsuto S, Yoshida S, Ueguchi-Tanaka M, et al. The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 2006;140(2):580–90.
Wang L, Xu YY, Ma QB, Li D, Xu ZH, Chong K. Heterotrimeric G protein alpha subunit is involved in rice brassinosteroid response. Cell Res. 2006;16(12):916–22.
Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, Qian Q, Zhu L, Chu C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009;58(5):803–16.
Tong H, Liu L, Jin Y, Du L, Yin Y, Qian Q, Zhu L, Chu C. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell. 2012;24(6):2562–77.
Jiang Y, Bao L, Jeong SY, Kim SK, Xu C, Li X, Zhang Q. XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice. Plant J. 2012;70(3):398–408.
Zhang C, Bai MY, Chong K. Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 2014;33(5):683–96.
Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, Uozu S, et al. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2002;32(4):495–508.
Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H. Matsuoka M. a rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell. 2003;15(12):2900–10.
Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. The Rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell. 2005;17(8):2243–54.
Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol. 2006;24(1):105–9.
Tanabe S, Ashikari M, Fujioka S, Takatsuto S, Yoshida S, Yano M, Yoshimura A, Kitano H, Matsuoka M, Fujisawa Y, et al. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell. 2005;17(3):776–90.
Mori M, Nomura T, Ooka H, Ishizaka M, Yokota T, Sugimoto K, Okabe K, Kajiwara H, Satoh K, Yamamoto K, et al. Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 2002;130(3):1152–61.
Liu X, Feng ZM, Zhou CL, Ren YK, Mou CL, Wu T, Yang CY, Liu SJ, Jiang L, Wan JM. Brassinosteroid (BR) biosynthetic gene lhdd10 controls late heading and plant height in rice (Oryza sativa L.). Plant Cell Rep. 2016;35(2):357–68.
Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10(2):231–43.
Ohnishi T, Watanabe B, Sakata K, Mizutani M. CYP724B2 and CYP90B3 function in the early C-22 hydroxylation steps of brassinosteroid biosynthetic pathway in tomato. Biosci Biotechnol Biochem. 2014;70(9):2071–80.
Wu C, Li X, Yuan W, Chen G, Kilian A, Li J, Xu C, Li X, Zhou DX, Wang S, et al. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 2003;35(3):418–27.
Liu E, Liu Y, Wu G, Zeng S, Tran Thi TG, Liang L, Liang Y, Dong Z, She D, et al. Identification of a candidate gene for panicle length in rice (Oryza sativa L.) via association and linkage analysis. Front Plant Sci. 2016;7:596.
Li M, Tang D, Wang K, Wu X, Lu L, Yu H, Gu M, Yan C, Cheng Z. Mutations in the F–box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice. Plant Biotechnol J. 2011;9(9):1002–13.
Ye H, Li L, Yin Y. Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J Integr Plant Biol. 2011;53(6):455–68.
Yang CJ, Zhang C, Lu YN, Jin JQ, Wang XL. The mechanisms of brassinosteroids’ action: from signal transduction to plant development. Mol Plant. 2011;4(4):588–600.
Duan K, Li L, Hu P, Xu SP, Xu ZH, Xue HW. A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J. 2006;47(4):519–31.
Li X, Gao X, Wei Y, Deng L, Ouyang Y, Chen G, Li X, Zhang Q, Wu C. Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5′-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell. 2011;23(4):1416–34.
Acknowledgements
We thank Ying Yang and Tuan Long for collecting panicle mutants.
Funding
This work was supported by the National Natural Science Foundation of China (31630054, 31425018, 31821005), Program for Chinese Outstanding Talents in Agricultural Scientific Research, and Program for Talent Plan of Wuhan Yellow Crane.
Availability of data and materials
All data used during the current study are included in this published article or are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
CW designed and supervised this research. YL and XL performed the main experiments. CW and YL wrote the manuscript. XL and DF performed the genetic analysis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Primers of SSR markers for mapping PMM1. (DOCX 16 kb)
Additional file 2:
Table S2. Predicted genes in the region containing PMM1. (DOCX 17 kb)
Additional file 3:
Figure S1. Sequence alignment of amino acids of OsDWARF4 and PMM1/OsDWARF11. (a) Sequence alignment of OsDWARF4 (CYP90B2) and OsDWARF11 (CYP724B1) from rice using the MEGA5.2 analysis tool. Identical and similar amino acid residues are shaded in black and gay, respectively. (b) Relative expression of OsDWARF4 in the root, stem, leaf and sheath and in developing panicles with 1, 2, 3.5, 4.5, 6.5, 8, 10 and 16.5-cm lengths before heading. Rice UBIQIUTIN5 was used as an internal control. Data are presented as means ± SE (n = 3). Significant at **P < 0.01. (DOCX 3903 kb)
Additional file 4:
Figure S2. Phenotype comparison of lamina joint inclination and plant height between the wild type (WT) and pmm1–1 plants. (a) A close-up view of flag leaf angles of the WT and pmm1–1 plants. (b) A close-up view of the secondary leaf angles the WT and pmm1–1 plants. (c) Measurements of flag leaf angles of the WT and pmm1–1 plants. (d) Measurements of the secondary leaf angles of WT and pmm1–1 plants. (e) The panicle and culm of the WT (left) and pmm1–1 (right). (f) The internodes and panicle of the WT (left) and pmm1–1 (right). (g) Measurements of the length between the WT and pmm1–1 plants. (h) The expression levels of genes associated with lamina joint inclination. (i) The expression levels of BR-related genes associated with internode elongation. Rice UBIQIUTIN5 was used as an internal control. Data are presented as means ± SE (n = 3). Significant at **P < 0.01. (DOCX 5072 kb)
Additional file 5:
Table S3. Primers for qRT-PCR, genotyping, and plasmid construction. (DOCX 18 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, Y., Li, X., Fu, D. et al. Panicle Morphology Mutant 1 (PMM1) determines the inflorescence architecture of rice by controlling brassinosteroid biosynthesis. BMC Plant Biol 18, 348 (2018). https://doi.org/10.1186/s12870-018-1577-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-018-1577-x