Abstract
Background
Rapeseed (Brassica napus L.) and soybean (Glycine max L.) seeds are rich in both protein and oil, which are major sources of biofuels and nutrition. Although the difference in seed oil content between soybean (~ 20%) and rapeseed (~ 40%) exists, little is known about its underlying molecular mechanism.
Results
An integrated omics analysis was performed in soybean, rapeseed, Arabidopsis (Arabidopsis thaliana L. Heynh), and sesame (Sesamum indicum L.), based on Arabidopsis acyl-lipid metabolism- and carbon metabolism-related genes. As a result, candidate genes and their transcription factors and microRNAs, along with phylogenetic analysis and co-expression network analysis of the PEPC gene family, were found to be largely associated with the difference between the two species. First, three soybean genes (Glyma.13G148600, Glyma.13G207900 and Glyma.12G122900) co-expressed with GmPEPC1 are specifically enriched during seed storage protein accumulation stages, while the expression of BnPEPC1 is putatively inhibited by bna-miR169, and two genes BnSTKA and BnCKII are co-expressed with BnPEPC1 and are specifically associated with plant circadian rhythm, which are related to seed oil biosynthesis. Then, in de novo fatty acid synthesis there are rapeseed-specific genes encoding subunits β-CT (BnaC05g37990D) and BCCP1 (BnaA03g06000D) of heterogeneous ACCase, which could interfere with synthesis rate, and β-CT is positively regulated by four transcription factors (BnaA01g37250D, BnaA02g26190D, BnaC01g01040D and BnaC07g21470D). In triglyceride synthesis, GmLPAAT2 is putatively inhibited by three miRNAs (gma-miR171, gma-miR1516 and gma-miR5775). Finally, in rapeseed there was evidence for the expansion of gene families, CALO, OBO and STERO, related to lipid storage, and the contraction of gene families, LOX, LAH and HSI2, related to oil degradation.
Conclusions
The molecular mechanisms associated with differences in seed oil content provide the basis for future breeding efforts to improve seed oil content.
Similar content being viewed by others
Background
Seed storage lipids not only provide food for human dietary consumption, but are also increasingly used as renewable sources for biofuels [1, 2]. In oil crops, such as Arabidopsis, soybean, rapeseed and sesame, seed oil content varies from 20 to 60%. Interestingly, the total seed storage reserves in soybean seed, consisting of ~ 20% oil and ~ 40% protein [3], is almost equal to the protein (~ 20%) and oil (~ 40%) contents in rapeseed [4]. As we know, most of the raw material required for seed oil and protein biosynthesis in rapeseed and soybean are derived from carbohydrate degradation [5]. And it should be noted that substrate competition between seed oil and protein synthesis exists in oilseed crops [6, 7]. This is because phosphoenolpyruvate (PEP), a carbon compound derived from glycolysis, is not only used to synthesize acetyl-Coenzyme A (acetyl-CoA), which serves as a substrate in the first step of de novo fatty acid synthesis, but is also required for the synthesis of oxaloacetate (OAA), which serves as a substrate in amino acid synthesis. Thus, carbon metabolism is related to oil synthesis, and boosting the carbon flow to lipid synthesis can significantly increase seed oil content [8].
In the past several decades, more than 700 acyl-lipid metabolism-related genes and several hundred genes participating in carbohydrate metabolism have been identified in Arabidopsis thaliana [9, 10]. Among these genes, more than 280 have been confirmed in A. thaliana mutants as associated with acyl-lipid metabolism (http://aralip.plantbiology.msu.edu) [11]. Meanwhile, many genes have been experimentally validated to be closely related to seed oil content. For example, phosphoenolpyruvate carboxylase (PEPC) in cotton [12], acetyl-CoA carboxylase (ACCase) in rapeseed [13] and potato [14] participate in de novo fatty acid biosynthesis; fatty acylthioesterase B (GmFatB) in soybean [15] and patatin-related phospholipase As (pPLAs) in Arabidopsis [16] are involved in fatty acid elongation; glycerol-3-phosphate dehydrogenase (GPDH) in rapeseed [17], glycerol-3-phosphate acyltransferase (GPAT) in Arabidopsis [18], 2-lysophosphatidic acid acyltransferase (LPAAT) in Arabidopsis [19] and cotton [20], acyl-CoA: diacylglycerol acyltransferase (DGAT) in Arabidopsis [21, 22], maize [23], and rapeseed [24] are related to TAG synthesis; oleosins (OLE1) in Arabidopsis [25] participates in lipid droplet assembly and storage. In addition, some transcription factors (TFs) have been found to be associated with seed oil content, i.e., WRINKLED1 (WRI1) [26], LEAFY COTYLEDON1 (LEC1) [27, 28], LEAFY COTYLEDON2 (LEC2) [29], FUSCA3 (FUS3) [30], GmDof4 and GmDof11 [31], GmbZIP123 [32], GmMYB73 [33], GmDREBL [34], GmNFYA [35], GmZF351 [36], and ABSCISIC ACID INSENSITIVE3 (ABI3) [37, 38]. However, all the above studies involved only a single lipid-related gene or transcription factor. Seed oil content is typically a quantitative trait regulated by multiple genes. As we know, these genes have been identified in the form of quantitative trait loci in soybean and rapeseed in the past decades [39,40,41,42,43].
In reality, acyl-lipid metabolism is a complex biological process that includes at least conversion of sucrose to pyruvate, plastidial de novo fatty acid (FA) synthesis, endoplasmic triacylglycerol (TAG) biosynthesis, and oil-body assembly. It is therefore important to determine whether specific combination of multiple genes from multiple metabolic pathways can increase seed oil content more effectively as compared with the manipulation of an individual gene. For example, it was found that Arabidopsis seed-specific overexpression of WRI1 and DGAT1 combined with suppression of SDP1 leads to higher seed oil content than the manipulation of each gene individually [44]. Additionally, the simultaneous overexpression of GmFabG (Glyma.12G092900), GmACP (Glyma.09G060900) and GmFAD8 (Glyma.03G056700) can significantly increase soybean seed oil content [45]. The down-regulation of CaFAD2 and CaFAE1 in crambe with the FAD2-FAE1 RNAi vector led to a significant increase in the seed oil to 80% compared to 13% for the wild type [46]. Seed-specific simultaneous overexpression of BnGPDH, BnGPAT and ScLPAAT genes in transgenic rapeseed may further enhance the desirable oil content relative to single-gene overexpression [47]. Moreover, Yu et al. [48] developed a complete analysis platform of functional annotation for the soybean genes involved in acyl-lipid metabolism, and this makes the study of acyl lipid metabolism more efficient and accurate. However, none of the above studies were conducted at the whole genome level.
With the rapid development of sequencing technology, more and more plant genomes have been sequenced, and this accelerates the progress of research on acyl-lipid metabolism [49,50,51,52,53]. Troncoso-Ponce et al. [10] showed that the expression stoichiometry of most key lipid-related genes was relatively conserved during seed development in Ricinus communis, Brassica napus, Euonymus alatus and Tropaeolum majus. Wang et al. [54] analyzed the expression differences of key ovule-specific genes between non-fibrous Raymond’s cotton and the upland cotton with fiber. Zhang et al. [55] dissected the molecular mechanisms of differences in seed oil content between four high-oil dicotyledons and three low-oil grass plants. However, little is known about the molecular mechanism for the difference of seed oil contents between soybean and rapeseed.
To understand the molecular mechanisms of the difference of seed oil content between rapeseed and soybean, an integrated omics analysis was performed in Arabidopsis, rapeseed, soybean and sesame based on Arabidopsis acyl-lipid metabolism- and carbon metabolism-related genes. The integrated omics analysis included gene copy number variation, expression pattern, microRNA/transcription factor, phylogenetic and co-expressional network analyses. Thus, candidate genes, transcription factors, and microRNAs that may be responsible for the difference were identified. These results provide a novel explanation for differences at the whole genome level and the basis for future breeding efforts to improve seed oil content in oilseed crops.
Results
Identification of candidate genes related to lipid biosynthesis
To identify orthologous genes in soybean and rapeseed, we used OrthoMCL to cluster putative OGs of genes across Arabidopsis, soybean, rapeseed and sesame. As a consequence, 172,626 (81.83%) protein-coding genes from the four species were clustered into 27,236 OGs (Additional file 1: Table S1), with each group representing a gene family. Among these gene families, 11,314 (41.54%) were defined as rapeseed-specific paralogous gene clusters without soybean genes, and only 3391 (12.45%) were soybean-specific families (Additional file 1: Table S2). In addition, 1687 OGs were identified to have one copy of a soybean gene and multiple copies for rapeseed genes, and 446 OGs with one copy for the rapeseed gene and multiple copies for soybean genes (Additional file 1: Table S2).
To date, more than 700 Arabidopsis acyl-lipid metabolism genes have been detected, of which 135 are directly involved in the processes of de novo fatty acid synthesis, triglyceride biosynthesis and lipid droplet formation. Using the method as described by Troncoso-Ponce et al. [10], we also extracted 238 Arabidopsis carbon-metabolism genes. Subsequently, searches using the 373 genes as queries were performed to obtain lipid biosynthesis-related homologous gene families (Additional file 1: Table S3). As a result, 230 putative OGs related to lipid synthesis were identified, which include 781 soybean genes and 1267 rapeseed genes (Additional file 1: Table S4).
Copy number variation and expression clustering of candidate genes related to lipid biosynthesis
In order to eliminate the effect of species ploidy, the relative copy number of a gene was used to measure the difference of copy number of homologous genes between species. Of the above 230 OGs related to lipid synthesis, 44 were found to have differences in relative copy number of a gene between soybean and rapeseed (Additional file 1: Table S5).
To cluster and visualize the expressional patterns of all 2048 rapeseed and soybean genes in the 230 OGs, we exploited STEM software [56] to analyze the expression data of four seed development stages in rapeseed (GSE77637, [57]) and soybean (GSE42871, [58]). In this study, R3, R4, R7 and R8 stages in soybean and 2, 4, 6, and 8 weeks after pollination (WAP) in rapeseed were defined as t1’, t2’, t3’ and t4’ in soybean and t1, t2, t3 and t4 in rapeseed, respectively. Results showed that 2048 genes were grouped into 20 clusters, including three up-regulation patterns (cluster13, cluster16 and cluster18) and one down-regulation profile (cluster3) during stages with rapid accumulation of seed oil (Additional file 2: Figure S1). Additionally, there were 23, 202, 86 and 22 genes in cluster3, cluster13, cluster16 and cluster18, respectively (P-value < 0.05) (Additional file 2: Figure S2).
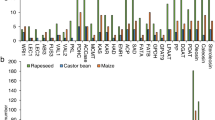
Based on the above two results, 192 soybean and 292 rapeseed candidate genes were inferred to be related to the differences of seed oil content between rapeseed and soybean. According to the sequence homology with Arabidopsis genes, 484 genes were found to putatively encode a series of core enzymes. For example, GRF2 and RBCS1A during photosynthesis; PGK, ApS1, SUC, PEPC and PKp involved in carbon metabolism from sucrose to pyruvate; PDK1, ACCase, KASII, HAD, KAR, FATA, SAD and FAD2 in de novo fatty acid biosynthesis; and PAP, PDCT participating in TAG synthesis, as well as oil-body proteins OBO, CALO, STERO, and oil degradation genes LOX, LAH, HSI2 and DSEL (Fig. 1).
Candidate genes for the difference of seed oil content between soybean and rapeseed. a Relative copy number variation analysis of candidate genes contributed to the difference of seed oil contents between soybean and rapeseed. The red genes in X-axis indicate more relative gene copies in rapeseed over in soybean, and the opposite situation is expressed in black. b PK in plastid is composed of Alpha (α) and Beta (β) subunits. ACCase contains homogeneous structure ACC2 and heterogeneous ACCase complex including α-CT, β-CT, BC, BCCP1 and BCCP2 subunits
To further determine the functions of the above 484 genes, KEGG enrichment analysis was conducted using KOBAS 2.0 [59]. As a result, the top 10 KEGG pathways for soybean and rapeseed candidate genes were obtained (Table 1). It was found that the two crops had eight KEGG pathways in common, namely pyruvate metabolism, carbon metabolism, biosynthesis of secondary metabolites, glycolysis, purine metabolism, biosynthesis of amino acids, carbon fixation in photosynthetic organisms, and glycerophospholipid metabolism. In addition, fatty acid biosynthesis in soybean is similar to fatty acid metabolism in rapeseed. The results are consistent with those in Troncoso-Ponce et al. [10] and Ohlrogge and Browse [60], and thus ensure the reliability of candidate genes in the next analysis.
Expression profiles of candidate genes responsible for the difference of seed oil content between rapeseed and soybean
The transcriptomic datasets from the seed developmental stages in soybean (GSE42871) and rapeseed (GSE77637) downloaded from the GEO (Gene Expression Omnibus) database were used to validate the above candidate genes. To compare the expressional profiles of each candidate gene in soybean and rapeseed, relative expression content for each gene was adopted in this study; this is defined as the ratio of the expression of each gene to average expression of all the genes in the species. As a result, a majority of candidate genes in rapeseed, except for phosphoenolpyruvate carboxylase (PEPC), Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (RBCS1A), lipoxygenase (LOX) and steroleosin (STERO), had higher relative expression than those in soybean, especially for PK and ACCase (Additional file 2: Figure S3, Additional file 1: Table S6).
More importantly, we noted some interesting phenomena. First, GmPEPC had higher relative expression at the early and middle seed development stages than BnPEPC (Fig. 3), indicating that PEP may be more likely to be used to synthesize protein in soybean seed, because PEPC, a member of carboxyl lyase family, catalyzes phosphoenolpyruvate (PEP) to produce oxaloacetic acid (OAA) for amino acid biosynthesis. Then, the relative expression contents of rapeseed genes encoding four subunits (α-CT, β-CT, BC and BCCP) of heterogeneous acetyl-CoA carboxylase (ACCase), catalyzing the first and committed reaction of de novo fatty acid biosynthesis in plastids, were higher than those of soybean genes in the oil rapid accumulation stages. Especially, BCCP1 and β-CT were not expressed during soybean seed development (Additional file 2: Figure S4). Finally, GmPEPC1 (Glyma.06G277500) and GmPEPC3 (Glyma.06G229900) had higher relative expression than PKp and ACCase in soybean. Conversely, PKp-β (BnaC02g44850D), PKp-α (BnaA01g24280D) and ACCase had higher relative expression than PEPC in rapeseed development stages (Additional file 2: Figure S4). Therefore, we deduced that PEPC, PKp and ACCase are most likely to be the key genes that regulate the distribution of carbon sources in soybean and rapeseed seeds.
Transcription factors and microRNAs regulatory network analysis of the candidate genes
To clarify the differences of regulatory networks of key candidate genes in soybean and rapeseed, we identified transcription factors (TFs) related to lipid biosynthesis in seed development. TFs and their target genes were downloaded from PlantTFDB v3.0 [61]. Together with the results mentioned above, it was found that the rapeseed-specific gene (BnaA10g13960D), encoding β-CT subunit of ACCase, is positively regulated by four zinc finger TFs, namely BnaA01g37250D, BnaA02g26190D, BnaC01g01040D and BnaC07g21470D (Table 2; Additional file 2: Figure S5) [62].
Meanwhile, 639 soybean and 92 rapeseed mature microRNAs were downloaded from miRBase (version 21). Their target genes were predicted using psRNATarget, a plant small RNA target analysis server. As a result, 4411 soybean and 1780 rapeseed miRNA-Target gene pairs were obtained. Among these genes, 116 and 61 were associated with lipid synthesis in soybean and rapeseed, respectively. Note that bna-miR169 inhibits the expression of the BnPEPC gene; this may facilitate a greater carbon flow to de novo fatty acid synthesis, and the expression of GmLPAAT2 gene is putatively inhibited by gma-miR171, gma-miR1516 and gma-miR5775 (Table 2; Additional file 2: Figure S5).
Evolutionary analysis of PEPC gene family
Phylogenetic analysis and conserved motifs analysis of PEPC gene family
Three plant-type PEPC genes (PTPCs) (AtPEPC1, AtPEPC2 and AtPEPC3) and one bacterial-type PEPC genes (BTPCs) (AtPEPC4) exist in A. thaliana [63]. To investigate the evolution of the PEPC gene family in soybean and rapeseed, the full amino acid sequences encoded by 33 PEPC genes in soybean, rapeseed and Arabidopsis were used to construct a phylogenetic tree using a neighbor-joining method. As a result, the PEPC genes were grouped into two distinct families with four subfamilies, which are consistent with those in A. thaliana (Fig. 2). In Fig. 2, GmPEPC1 is close to GmPEPC3 in its evolutionary relationship, and their expression patterns during seed development in Additional file 2: Figure S4 are complementary. Meanwhile, the conserved motifs of PEPC genes were further analyzed using MEME [64]. Results showed that the motif structure of GmPEPC3 was more conserved than that of BnPEPC3, and the motif structures of BnPEPC1 and BnPEPC2 were also relatively more conserved than that of BnPEPC3 (Fig. 2). Note that there are distinct differences of BTPCs between soybean and rapeseed, indicating the existence of its extensive functional differentiation.
Phylogenetic tree and gene motif analysis of PEPC gene family. a Phylogenetic tree of PEPC gene family is constructed from the complete alignment of 33 PEPC protein sequences in Arabidopsis, soybean, and rapeseed using the neighbor-joining method with 1000 bootstrap replicates with the MEGA 7.0 software program. The bootstrap scores are indicated on the nodes, and the 4 PEPC branches, all of which are based on Arabidopsis PEPC orthologous genes, are indicated in four color boxes. The representative gene of each branch is shown followed by an additional abbreviation. b Conserved domains analysis of PEPCs. The domains of soybean genes in AtPEPC3 branch are relatively more conservative compared with rapeseed genes. And the domains of rapeseed genes in AtPEPC1 and AtPEPC2 branches are relatively conservative. However, the differences of gene domain in the bacterial AtPEPC4 branch are significant between soybean and rapeseed
Evolutionary rate and positive selection analysis of PEPC genes
To determine whether the genes of the PEPC gene family are under different evolutionary constraints in soybean and rapeseed, the ω (Ka/Ks) values for the above genes were calculated using the branch model (BM) and the branch-site model (BSM) of the Codeml program in PAML. As a result, the evolution rate ω0 (Ka/Ks) was estimated to be 0.340 and log-likelihood was − 2343.655 if the evolution rates at all branches were the same; six evolution rates (ω) were estimated to be 0.078, 0.054, 0.106, 0.093, 0.495 and 0.175 and log-likelihood was − 2361.163 if the evolution rates changed across different branches (Additional file 3: Table S7; Additional file 2: Figure S6). Clearly, there was a significant difference between the above two models (Additional file 3: Table S7, P-value = 4.278e-06). We also found that the ω value for the bacterial-type PEPC soybean sub-branch (ω = 0.495) was significantly higher than the ω0 value (= 0.340) and the ω values for the other sub-branches were significantly lower than the ω0 value (= 0.340). This indicates that the BTPC genes have experienced positive selection and the PTPC genes have experienced purifying selection. Furthermore, the BSM model was used to identify positively selected sites. As a result, we observed significant positive selection for soybean bacterial-type PEPC genes and the amino acid sites of positive selection, associated with GmPEPC4, were 56F and 61 V (Additional file 3: Table S8).
Differential analysis of PEPC1 gene co-expression networks between soybean and rapeseed
The co-expression network of one gene is frequently constructed by Pearson’s correlation coefficient [65, 66]. In the present study this method was used to construct the co-expression networks of the PEPC1 gene in soybean and rapeseed. The differences between the two networks were also used to identify extra candidate genes. As a result, 121 soybean and 133 rapeseed genes were co-expressed with GmPEPC1 and BnPEPC1, respectively. Among these co-expressed genes, 17 were orthologous. The other genes were used to conduct KEGG pathway enrichment analysis. In the top 10 KEGG pathways for soybean or rapeseed, the soybean-specific biological process is “the synthesis of valine, leucine and isoleucine”, involving Glyma.13G148600, Glyma.13G207900 and Glyma.12G122900, and rapeseed-specific biological process is “plant circadian rhythms”, involving genes BnSTKA (BnaC08g48660D, BnaA09g42220D, BnaA01g21040D, BnaC08g34660D, BnaC01g42660D) and BnCKII (BnaC08g30500D, BnaC02g33100D, BnaC04g05080D, BnaA02g24960D) (Fig. 3).
Comparison of PEPC1 gene co-expression networks in rapeseed (a) and soybean (b). 17 genes in light pink are orthologous genes in rapeseed and soybean. More than 100 genes in red were enriched in the same processes based on KEGG pathway enrichment analysis. The blue nodes represent rapeseed genes enriched-specific in “plant circadian rhythms” (a) and soybean genes enriched-specific in “Valine, leucine and isoleucine biosynthesis” (b), respectively
Discussion
PEPC, along with its miRNA and co-expressed genes, which can affect the flow of carbon sources in seeds, may contribute to the difference of seed oil content between soybean and rapeseed
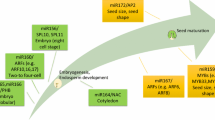
Seed oil content is almost negatively correlated to seed protein content in soybean and rapeseed [67,68,69]. As we know, PEP is used to synthesize acetyl-CoA under the catalysis of pyruvate kinase (PK) and acetyl coenzyme A carboxylase (ACCase) so that the PEP enters into the fatty acid synthesis pathway. Additionally, PEP is also used to synthesize oxaloacetate (OAA) under the catalysis of phosphoenolpyruvate carboxylase (PEPC) so that the PEP enters into the amino acid synthesis pathway. Results in this study showed that PEPC genes, together with their miRNA and co-expressed genes, may increase the flow of carbon to the biosynthesis of amino acids in soybean seed and to the de novo fatty acid synthesis in rapeseed seed, resulting in the difference of seed oil content between the two species (Fig. 4). The reasons are as follows.
Molecular mechanisms for the difference of seed oil content between soybean and rapeseed. Candidate genes contributed to the differences of seed oil content between soybean and rapeseed obtained in the study were marked with red color. GRF2 and RBCS1A: photosynthesis; PGK, ApS1, SUC, PEPC and PKp: carbon metabolism from sucrose to pyruvate; PDK1, ACCase, KASII, HAD, KAR, FATA, SAD and FAD2: in de novo fatty acid biosynthesis; PAP and PDCT: TAG synthesis; OBO, CALO and STERO: oil-body protein genes; LOX, LAH, HSI2 and DSEL: oil degradation genes. Among these candidate genes, BCCP1 (BnaA03g06000D) and β-CT (BnaC05g37990D) in heterogeneous acetyl-CoA carboxylase (ACCase) are rapeseed-specific genes, and β-CT is positively regulated by four transcription factors (BnaA01g37250D, BnaA02g26190D, BnaC01g01040D and BnaC07g21470D). The gene expression of PEPC1 in rapeseed is putatively inhibited by bna-miR169, while LPAAT in soybean putatively inhibited by gma-miR171, gma-miR1516 and gma-miR5775 in triglyceride synthesis. The pink genes are speculated related specifically to high seed oil content of rapeseeds, and the purple speculated specifically related to high seed protein content in soybean, which were both identified by PEPC co-expression network analysis. Soybean genes participated in Branched-Chain Amino Acid (BCAA) synthesis may contribute to seed high protein content by adjusting the flow of PEP and downstream protein biosynthesis. Rapeseed genes BnSTKA and BnCKII are likely to promote the triglyceride synthesis by phosphorylating circadian TFs cca1/lhy and thus increase the seed oil content of rapeseed
First, GmPEPC1 has higher relative expression at the early and middle stages of seed development than BnPEPC1, and bna-miR169 putatively inhibits the expression of BnPEPC. Although the bacterial-type PEPC (BTPC) gene in A. thaliana can inhibit the expression of the plant-type PEPC (PTPC) gene [63, 70,71,72,73], BTPC genes in soybean have experienced positive selection (Additional file 3: Tables S7 and S8) and it is possible to lose the function of inhibiting the expression of PTPC gene (GmPEPC1). In addition, Xu et al. [12] increased the accumulation of cotton seed oil by the down-regulation of GhPEPC1 via RNA interference in transgenic cotton plants. These studies provide evidence for greater carbon flow to amino acid metabolism in soybean seed and to de novo fatty acid synthesis in rapeseed seed. This may partly explain why there are high seed protein content in soybean and high seed oil content in rapeseed.
Secondly, the expression of LPAAT2-encoding gene, involved in triacylglycerol synthesis in soybean seed, is putatively inhibited by three miRNAs (gma-miR171, gma-miR1516 and gma-miR5775) based on the results of bioinformatics analysis (Table 2).
Finally, gene co-expression network analysis helps us to understand different biological pathways in soybean and rapeseed. KEGG enrichment analyses of genes co-expressed with GmPEPC1 showed that three soybean genes (Glyma.13G148600, Glyma.13G207900 and Glyma.12G122900) were enrich-specific in the “leucine, isoleucine and valine” synthesis pathway. Leucine, isoleucine and valine are the three major branched-chain amino acids for protein synthesis. The content of branched-chain amino acids in seeds is positively correlated with the protein content in general, which can effectively maintain the accumulation of storage proteins in seeds [74]. Therefore, it is speculated that the expression of such soybean genes may be beneficial for the accumulation of storage proteins in seeds. Meanwhile, KEGG enrichment analysis of genes co-expressed with BnPEPC1 revealed that nine rapeseed genes, encoding serine/threonine-protein kinase (BnSKTA; BnaC08g48660D, BnaA09g42220D, BnaA01g21040D, BnaC08g34660D and BnaC01g42660D) and Casein kinase II subunit beta (BnCKII; BnaC08g30500D, BnaC02g33100D, BnaC04g05080D and BnaA02g24960D), were specifically enriched in the “plant circadian rhythm” category, which can regulate seed oil metabolism and hormone signaling pathway [75, 76]. Lipid metabolism is subject to diurnal regulation at the early stages of seed development in Arabidopsis [77]; diurnal differential expression of genes encoding JcDof1, a dof TF of Jatropha curcas in response to light signal, β-hydroxy-3-methylglutaryl-CoA reductase and Cyp7A1, regulates seed oil synthesis and accumulation [78, 79]. CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) from the core clock system can affect the reserve mobilization of storage lipid [77], but this process is affected by the phosphorylation of protein kinase (CK2) [80]. Therefore, the phosphorylation of genes BnSTKA and BnCKII may promote the storage of seed oil.
Rapeseed-specific genes encoding β-CT and BCCP1 subunits of acetyl-CoA carboxylase and transcription factors may be associated with higher seed oil content in rapeseed
β-CT and BCCP are important components for heterogeneous acetyl-CoA carboxylase (ACCase) [81, 82]. Overexpression of ACCase subunit genes can significantly increase fatty acid content in oil crop seed [13, 14, 83]. In this study, soybean β-CT and BCCP1 was not expressed, while BCCP2, BC and α-CT showed high expression during seed development stages (Additional file 2: Figure S4). This is consistent with the results of Zhang et al. [55]. Meanwhile, genes BCCP1, BCCP2, BC, α-CT and β-CT showed high expression during rapeseed seed development stages. Especially, the β-CT subunit gene (BnaA10g13960D) was positively regulated by four zinc finger C2H2 transcription factors (BnaA01g37250D, BnaA02g26190D, BnaC01g01040D and BnaC07g21470D) predictably, which is consistent with the results of Jin et al. [62]. Similarly, Li et al. [36] demonstrated that overexpression of GmZF351, a gene encoding a tandem CCCH zinc finger protein, can activate lipid biosynthesis genes and increase seed oil accumulation in soybean. Moreover, Li et al. [84] found that transfer DNA insertional alleles that completely eliminate the accumulation of BCCP2 have no perceptible effect on fatty acid accumulation, while reducing the BCCP1 accumulation can dramatically decreases fatty acid accumulation in Arabidopsis thaliana. This partly supports that rapeseed-specific BCCP1 gene may associate with high seed oil content of rapeseed. It should also note that RNA levels don’t always equate to protein and/or lipid metabolite levels in plants [85].
The expansion of gene families associated with lipid storage and the contraction of gene families related to lipid degradation may contribute to high seed oil content in rapeseed
Seed triglyceride is mainly stored in lipid droplets, and the size of the lipid droplets and the spatial distribution of their assembly proteins affect seed oil content [86, 87]. In this study, it was found that the relative copy numbers of genes encoding STERO, CALO and OLEs in rapeseed are significantly higher than those in soybean (Fig. 1, Additional file 1: Table S6), and such genes in rapeseed are obviously up-regulated during stages of rapid lipid accumulation (t2~t3) (Additional file 2: Figure S3, Additional file 1: Table S6). On the other hand, the gene families LOX, LAH and HSI2, related to lipid degradation, have contracted in rapeseed. In other words, the relative copy numbers of these genes are much smaller than those in soybean (31/4 < 41/2, 10/4 < 24/2 and 8/4 < 8/2, (gene absolute copy numbers) / (species polyploidy)). This relationship was also found between soybean and sesame. Specifically, this latter species shows contraction of gene families (LOX, LAH and FAR1) related to lipid degradation and expansion of gene families (LTP1 and SUT) related to lipid storage [51]. Therefore, it was speculated that the contraction of gene families related to lipid degradation and the expansion of gene families related to lipid storage may be an important reason for the higher seed oil content in rapeseed than in soybean.
In order to further ascertain whether the degradation of seed storage materials in oilseed crop is specialized in an evolutionary sense, we investigated gene families related to protein degradation in soybean, rapeseed and sesame seeds. As we know, the degradation of protein in plant cells is mainly mediated by the ubiquitin proteasome, lysosomal and caspase pathways. Among the three pathways, the ubiquitination proteasome pathway is the main pathway of storage protein degradation in oilseed crop seeds [88], and it mainly involves ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin ligases (E3) [89]. In this study, we found that the relative copy numbers of genes encoding E1, E2 and E3 in soybean, rapeseed and sesame were 4/2 = 8/4 > 2/2, 44/2 < 105/4 > 23/2 and 33/2 > 52/4 > 10/2, which are not consistent with the protein contents in soybean, rapeseed and sesame seeds (~ 40%, ~ 20%, and ~ 17%). This indicates that the phenomenon, which has a bias to consume protein or oil mainly to power the life activities during seed development, does not occur in the evolution of oil crops.
More evidence for candidate genes that are associated with the seed oil content difference between soybean and rapeseed
Many candidate genes predicted in this study could be responsible for the difference of seed oil content between soybean and rapeseed have been experimentally confirmed to be related to seed oil content. In addition to those mentioned above, with the up-regulated expression for the genes BnGRF2 and BnRBCS1A and the down-regulated expression of gene BnPDK1, Hu et al. [69] cultivated a rapeseed line YN171 with a super high seed oil content of 64.8% (Fig. 1). Similarly, with the down-regulated expression of gene GmFAD2–1 by RNA interference, seed oleic acid content in soybean increased to 94.58% and the linoleic acid content decreased to < 3% (Fig. 1) [15].
Differentially expressed genes, associated with seed oil content and identified among cultivars with different seed oil content, also provide relevant evidence. Among the 33 differentially expressed genes identified in rapeseed by Xu et al. [90], PDAT (Additional file 1: Table S4) and OBO (Fig. 1) are also found in the present study. Among the 28 core enzymes involved in lipid synthesis in soybean [55], 8 were also found in the present study (Additional file 3: Table S9).
In this study, we are focusing on the difference of total seed oil content between soybean and rapeseed. However, their other differences exist as well, i.e., seed oil composition, grown climatic environment, nitrogen fixation, and species characteristics, which likely affect the conclusion in this study.
Conclusion
In this study, we identified candidate genes and their transcription factors and microRNAs to explain the difference in seed oil content between soybean and rapeseed. First, PEPC, along with its microRNAs and co-expression genes, affect the carbon source flow in seeds, which may lead to differences in seeds oil content. Then, BCCP1 and β-CT and its transcription factors that are characteristic of rapeseed may result in high seeds oil content. Finally, the expansion of gene families related to lipid storage, and the contraction of gene families related to oil degradation may play important roles on the difference in seed oil content.
Methods
Data sources
Sequences were collected using the similar method described by Tatusov et al. [91]. Protein-coding transcripts of Arabidopsis (TAIR release 10, https://www.arabidopsis.org/), rapeseed (release 4.1, http://www.genoscope.cns.fr/brassicanapus/), soybean (release Wm82.a2.v1, https://www.soybase.org/) and sesame (release 1.0, http://ocri-genomics.org/Sinbase/) were downloaded, respectively. If a gene has multiple transcripts, the longest was selected.
The transcriptome data of soybean (G. max Williams 82) [58] (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42871) and rapeseed (B. napus Darmor-bzh) [57] (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE77637. ) were downloaded from Gene Expression Omnibus (GEO). Rapeseed transcriptome data included four seed developmental stages: 2, 4, 6, and 8 weeks after pollination (WAP), and soybean transcriptome data included seven seed developmental stages: whole seed 4 days after fertilization (DAF), whole seed 12–14 DAF, whole seed 22–24 DAF, whole seed 5–6 mg in weight, cotyledons 100–200 mg in weight, cotyledon 400–500 mg in weight, and dry whole seed. Of which, whole seed 12–14 DAF, whole seed 22–24 DAF, cotyledon 400–500 mg in weight, and dry whole seed in soybean are almost respectively equal to 2, 4, 6, and 8 DAF based on the definition of soybean vegetative and reproductive growth [92], which are consistent with 2, 4, 6, and 8 WAP in rapeseed. Thus, we selected the four stages of soybean and rapeseed seed development mentioned above for subsequent analysis. The genes expression level (RPKM: reads per kilobase per million mapped reads) were normalized and quantified by the DESeq package in Bioconductor [93].
Delimitation of orthologous genes
Identification of orthologous groups (OGs) in Arabidopsis, soybean, rapeseed and sesame was conducted using OrthoMCL software with default parameters [94]. Based on all-against-all BLASTP of non-redundant protein sequences, clusters were obtained according to reciprocal best similarity pairs between and within species, using OrthoMCL software implemented by the Markov clustering algorithm (MCL; http://micans.org/mcl/) [95]. To obtain more accurate results, two other known methods, namely Proteinortho [96] and Inparanoid 8 [97], were also used to determinate OGs of soybean and rapeseed.
Prediction of candidate genes related to carbon metabolism and lipid biosynthesis
Acyl-lipid biosynthesis process is mainly involved in fatty acid synthesis and elongation from pyruvate, TAG synthesis, and oil-body storage. In Arabidopsis, 135 acyl-lipid biosynthesis-related genes were downloaded from ARALIP (http://aralip.plantbiology.msu.edu/) [9], and 238 carbon metabolism-related genes were also obtained using the method of Troncoso-Ponce et al. [10] with a slight modification. Such genes were used as query, along with OGs, to identify carbon metabolism- and lipid biosynthesis-related OGs. To further identify candidate genes for the differences of seed oil content, gene expression pattern clustering and interspecific relative copy numbers variation analysis were carried out. Gene expression clustering analysis was performed using Short Time-series Expression Miner (STEM, http://www.cs.cmu.edu/~jernst/stem/) [56] with the following parameters: Log normalize a time series vector of gene expression values (v0, v1, v2, ..., vn) to (0, log2(v1/v0), log2(v2/v0), ⋯, log2(vn/v0)), Minimum Absolute Expression Change 2, −p 0.05.
KEGG pathway enrichment analysis
Kyoto Encyclopedia of Gene and Genome (KEGG) pathway enrichment analysis was performed using the online tool KOBAS (version 2.0; http://kobas.cbi.pku.edu.cn/index.php) [59]. The P-values for each KEGG biological process was calculated by Fisher’s exact test [59]. To control the false discovery rate (FDR ≤ 0.05), the Benjamini-Hochberg method was used to conduct multiple testing correction [98]. In addition, the small term cutoff value was set at 5.
Phylogenetic analysis and motif analysis
The full-length amino acid sequence alignments were performed using MUSCLE [99] with default parameters and then phylogenetic tree reconstruction was conducted with both Neighbor Joining (NJ) and Maximum Likelihood (ML) approaches in MEGA 7.0 [100]. In the NJ method, parameter setups were as follows: - model: poisson correction; Bootstrap: 1000 replicates; and gap/missing data: pairwise deletion. To ensure the accurateness of ML tree, which is constructed to eliminate the long-branch attraction (LBA) caused by distant species, we also used maximum likelihood approaches with PhyML v3.0 [101], and estimated the best-fitting models with the jModeltest software [102]. The phylogenetic tree was displayed, annotated and managed using iTOL (https://itol.embl.de/) [103]. Conserved functional motifs were identified using the program Multiple Em for Motif Elicitation [64] (MEME v4.11.2, http://meme-suite.org/tools/meme) with the following parameters: - the width of a motif was between 6aa and 50aa, and the number of motifs was no more than 20.
Selective pressure and positive selection analyses
The amino acid sequences were aligned using MUSCLE [99], alignment gaps were manually deleted, and then used for following calculations. The ratio (ω value) of nonsynonymous substitution rate (Ka) to synonymous substitution rate (Ks) of homologous gene pairs was computed with the maximum likelihood method of the branch model in Codeml from the PAML package (version 4.9) [104].
To test for the variation of the ω ratio among different branches in gene trees, a branch-specific model was used and conducted in Codeml. The branch-specific model allows the ω ratio to vary among branches in the phylogeny (model = 2, NSsites = 0), and it could be used to test whether there are different ω values on particular lineages [105]; thus, this model can be compared with the one-ratio model (model = 0, NSsites = 0) that assumes a constant ω value across all branches using the likelihood ratio test (LRT). The datasets used in Ka/Ks ratio estimation were further used in the next positive selection analysis of the branch-site model (BSM) using the Bayes empirical Bayes method described by Yang et al. [104].
Transcription factor (TF)- and microRNA-targets analysis
Soybean and rapeseed microRNAs were downloaded from miRBase (release 21, http://www.mirbase.org/) [106]. psRNATarget (http://plantgrn.noble.org/psRNATarget) [107] was used to identify miRNA targets with default parameters except for the Expectation (e) and Max UPE, which were set at 3 and 25, respectively. The transcription factors (TFs) and TF-target pairs were downloaded directly from PlantTFDB 3.0 (http://planttfdb.cbi.pku.edu.cn/) [61]. To ascertain whether miRNAs controls target-genes expression in seed development stages, bioinformatic analysis software miRDB (http://mirdb.org/miRDB/) [108] was used to preliminarily verify whether there is a putative binding site for miRNAs in the 3′-UTR of target-genes mRNA.
Gene co-expression network analysis
Pearson’s correlation coefficients (r) were calculated using the ‘cor’ function of R package. The gene expression data (Reads Per Kilobase per Million mapped reads: RPKM) was used to calculate the correlation coefficients between genes. The criteria for determining co-expressional genes were set at r ≥ 0.9 or r ≤ − 0.9 and P-values ≤ 0.05 [65]. Graphical visualization of the gene co-expression network was performed using Cytoscape 3.4.0 (http://www.cytoscape.org/) [109]. Genes co-expressed with the target gene, meeting the filter criteria, were further used to conduct KEGG pathway enrichment analysis using KOBAS 2.0 [59].
Abbreviations
- ACCase:
-
Acetyl CoA carboxylase
- ApS1:
-
ADP-glucose pyrophosphorylase
- CALO:
-
Caleosin
- DSEL:
-
Phospholipase
- FAD:
-
Oleoyl desaturase
- FATA:
-
Fatty acylthioesterase A
- GEO:
-
Gene Expression Omnibus
- GRF2:
-
Growth-regulating factor 2
- HAD:
-
Hydroxyacyl-ACP Dehydrase
- HSI2:
-
High-level expression of sugar-inducible gene 2
- KAR:
-
Ketoacyl-ACP reductase
- KASII:
-
Ketoacyl-ACP Synthase II
- LAH:
-
Lipid Acylhydrolase-like
- LBA:
-
Long-branch attraction
- LOX:
-
Lipoxygenase
- LPAAT:
-
Lysophosphatidic acid acyltransferase
- MCL:
-
Markov clustering algorithm
- OAA:
-
Oxalacetic acid
- OBO:
-
Oil-Body Oleosin
- OGs:
-
Orthologous groups
- PAP:
-
Phosphatidate phosphatase
- PDCT:
-
Phosphatidylcholine:diacylglycerol cholinephosphotransferase
- PDK1:
-
Pyruvate dehydrogenase kinase isozyme 1
- PEP:
-
Phosphoenolpyruvate
- PEPC:
-
Phosphoenolpyruvate carboxylase
- PGK:
-
Phosphoglycerate kinase
- PK:
-
Pyruvate kinase
- PKp:
-
Plastidial pyruvate kinase
- PYR:
-
Pyruvic acid
- RBCS1A:
-
Ribulose bisphosphate carboxylase small chain 1A
- SAD:
-
Stearyl-ACPdesaturase
- STERO:
-
Steroleosin
- SUC:
-
Sucrose transport protein
- WAP:
-
Weeks after pollination
References
Durrett TP, Benning C, Ohlrogge J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 2008;54(4):593–607.
Horn PJ, Benning C. The plant lipidome in human and environmental health. Science. 2016;353(6305):1228–32.
Min CW, Gupta R, Kim SW, Lee SE, Kim YC, Bae DW, Han WY, Lee BW, Ko JM, Agrawal GK, Rakwal R, Kim ST. Comparative biochemical and proteomic analyses of soybean seed cultivars differing in protein and oil contents. J Agric Food Chem. 2015;63(32):7134–42.
Kennedy Y, Yokoi S, Sato T, Daimon H, Nishida I, Takahata Y. Genetic variation of storage compounds and seed weight in rapeseed (Brassica napus L.) germplasms. Breeding Sci. 2011;61(3):311–5.
Rawsthorne S. 2002 Carbon flux and fatty acid synthesis in plants. Prog Lipid Res. 2002;41(2):182–96.
Voelker T, Kinney AJ. Variations in the biosynthesis of seed-storage lipids. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:335–61.
Thelen JJ, Ohlrogge JB. Metabolic engineering of fatty acid biosynthesis in plants. Metab Eng. 2002;4(1):12–21.
Weselake RJ, Taylor DC, Rahman MH, Shah S, Laroche A, McVetty PB, Harwood JL. Increasing the flow of carbon into seed oil. Biotechnol Adv. 2009;27(6):866–78.
Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, Franke RB, Graham IA, Katayama K, Kelly AA, Larson T, Markham JE, Miquel M, Molina I, Nishida I, Rowland O, Samuels L, Schmid KM, Wada H, Welti R, Xu C, Zallot R, Ohlrogge J. Acyl-Lipid Metabolism. In: Rockville ed., the Arabidopsis book. MD: American Society of Plant Biologists. 2013;11:e0161.
Troncoso-Ponce MA, Kilaru A, Cao X, Durrett TP, Fan J, Jensen JK, Thrower NA, Pauly M, Wilkerson C, Ohlrogge JB. Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 2011;68(6):1014–27.
McGlew K, Shaw V, Zhang M, Kim RJ, Yang W, Shorrosh B, Suh MC, Ohlrogge J. An annotated database of Arabidopsis mutants of acyl lipid metabolism. Plant Cell Rep. 2015;34(4):519–32.
Xu Z, Li J, Guo X, Jin S, Zhang X. Metabolic engineering of cottonseed oil biosynthesis pathway via RNA interference. Sci Rep. 2016;13(6):33342.
Roesler K, Shintani D, Savage L, Boddupalli S, Ohlrogge J. Targeting of the Arabidopsis homomeric acetyl-coenzyme a carboxylase to plastids of rapeseeds. Plant Physiol. 1997;113(1):75–81.
Klaus D, Ohlrogge JB, Neuhaus HE, Dörmann P. Increased fatty acid production in potato by engineering of acetyl-CoA carboxylase. Planta. 2004;219(3):389–96.
Murad AM, Vianna GR, Machado AM, da Cunha NB, Coelho CM, Lacerda VA, Coelho MC, Rech EL. Mass spectrometry characterisation of fatty acids from metabolically engineered soybean seeds. Anal Bioanal Chem. 2014;406:2873–83.
Li M, Bahn SC, Fan C, Li J, Phan T, Ortiz M, Roth MR, Welti R, Jaworski J, Wang X. Patatin-related phospholipase pPLAIIIδ increases seed oil content with long-chain fatty acids in Arabidopsis. Plant Physiol. 2013;162(1):39–51.
Vigeolas H, Waldeck P, Zank T, Geigenberger P. Increasing seed oil content in oil-seed rape (Brassica napus L.) by over-expression of a yeast glycerol-3-phosphate dehydrogenase under the control of a seed-specific promoter. Plant Biotechnol J. 2007;5(3):431–41.
Jain RK, Coffey M, Lai K, Kumar A, MacKenzie SL. Enhancement of seed oil content by expression of glycerol-3-phosphate acyltransferase genes. Biochem Soc Trans. 2000;28(6):958–61.
Maisonneuve S, Bessoule JJ, Lessire R, Delseny M, Roscoe TJ. Expression of rapeseed microsomal lysophosphatidic acid acyltransferase isozymes enhances seed oil content in Arabidopsis. Plant Physiol. 2010;152(2):670–84.
Wang N, Ma J, Pei W, Wu M, Li H, Li X, Yu S, Zhang J, Yu J. A genome-wide analysis of the lysophosphatidate acyltransferase (LPAAT) gene family in cotton: organization, expression, sequence variation, and association with seed oil content and fiber quality. BMC Genomics. 2017;18(1):218.
Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, Covello PS, Taylor DC. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 2001;126(2):861–74.
Misra A, Khan K, Niranjan A, Nath P, Sane VA. Over-expression of JcDGAT1 from Jatropha curcas increases seed oil levels and alters oil quality in transgenic Arabidopsis thaliana. Phytochemistry. 2013;96:37–45.
Zheng P, Allen WB, Roesler K, Williams ME, Zhang S, Li J, Glassman K, Ranch J, Nubel D, Solawetz W, Bhattramakki D, Llaca V, Deschamps S, Zhong GY, Tarczynski MC, Shen B. A phenylalanine in DGAT is a key determinant of oil contents and composition in maize. Nat Genet. 2008;40(3):367–72.
Weselake RJ, Shah S, Tang M, Quant PA, Snyder CL, Furukawa-Stoffer TL, Zhu W, Taylor DC, Zou J, Kumar A, Hall L, Laroche A, Rakow G, Raney P, Moloney MM, Harwood JL. Metabolic control analysis is helpful for informed genetic manipulation of oilseed rape (Brassica napus) to increase seed oil content. J Exp Bot. 2008;59(13):3543–9.
Shimada TL, Hara-Nishimura I. Oil-body-membrane proteins and their physiological functions in plants. Biol Pharm Bull. 2010;33(3):360–3.
Ma W, Kong Q, Grix M, Mantyla JJ, Yang Y, Benning C, Ohlrogge JB. Deletion of a C-terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J. 2015;83(5):864–74.
Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang XJ, Zuo J. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008;148(2):1042–54.
Tan H, Yang X, Zhang F, Zheng X, Qu C, Mu J, Fu F, Li J, Guan R, Zhang H, Wang G, Zuo J. Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 2011;156(3):1577–88.
Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007;50(5):825–38.
Wang H, Guo J, Lambert KN, Lin Y. Developmental control of Arabidopsis seed oil biosynthesis. Planta. 2007;226(3):773–83.
Wang HW, Zhang B, Hao YJ, Huang J, Tian AG, Liao Y, Zhang JS, Chen SY. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 2007;52(4):716–29.
Song QX, Li QT, Liu YF, Zhang FX, Ma B, Zhang WK, Man WQ, Du WG, Wang GD, Chen SY, Zhang JS. Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. J Exp Bot. 2013;64(14):4329–41.
Liu YF, Li QT, Lu X, Song QX, Lam SM, Zhang WK, Ma B, Lin Q, Man WQ, Du WG, Shui GH, Chen SY, Zhang JS. Soybean GmMYB73 promotes lipid accumulation in transgenic plants. BMC Plant Biol. 2014;14:73.
Zhang YQ, Lu X, Zhao FY, Li QT, Niu SL, Wei W, Zhang WK, Ma B, Chen SY, Zhang JS. Soybean GmDREBL increases lipid content in seeds of transgenic Arabidopsis. Sci Rep. 2016;6:34307.
Lu X, Li QT, Xiong Q, Li W, Bi YD, Lai YC, Liu XL, Man WQ, Zhang WK, Ma B, Chen SY, Zhang JS. The transcriptomic signature of developing soybean seeds reveals genetic basis of seed trait adaptation during domestication. Plant J. 2016;86(6):530–44.
Li QT, Lu X, Song QX, Chen HW, Wei W, Tao JJ, Bian XH, Shen M, Ma B, Zhang WK, Bi YD, Li W, Lai YC, Lam SM, Shui GH, Chen SY, Zhang JS. Selection for a zinc-finger protein contributes to seed oil increase during soybean domestication. Plant Physiol. 2017;173(4):2208–24.
Crowe AJ, Abenes M, Plant A, Moloney MM. The seed-specific transactivator, ABI3, induces oleosin gene expression. Plant Sci. 2000;151(2):171–81.
Mönke G, Seifert M, Keilwagen J, Mohr M, Grosse I, Hähnel U, Junker A, Weisshaar B, Conrad U, Bäumlein H, Altschmied L. Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res. 2012;40(17):8240–54.
Jiang C, Shi J, Li R, Long Y, Wang H, Li D, Zhao J, Meng J. Quantitative trait loci that control the oil content variation of rapeseed (Brassica napus L.). Theor Appl Genet. 2014;127(4):957–68.
Sun F, Liu J, Hua W, Sun X, Wang X, Wang H. Identification of stable QTLs for seed oil content by combined linkage and association mapping in Brassica napus. Plant Sci. 2016;252:388–99.
Sun M, Hua W, Liu J, Huang S, Wang X, Liu G, Wang H. Design of new genome- and gene-sourced primers and identification of QTL for seed oil content in a specially high-oil Brassica napus cultivar. PLoS One. 2012;7(10):e47037.
Hwang EY, Song Q, Jia G, Specht JE, Hyten DL, Costa J, Cregan PB. A genome-wide association study of seed protein and oil content in soybean. BMC Genomics. 2014;15:1.
Cao Y, Li S, Wang Z, Chang F, Kong J, Gai J, Zhao T. Identification of major quantitative trait loci for seed oil content in soybeans by combining linkage and genome-wide association mapping. Front Plant Sci. 2017;8:1222.
van Erp H, Kelly AA, Menard G, Eastmond PJ. Multigene engineering of triacylglycerol metabolism boosts seed oil content in Arabidopsis. Plant Physiol. 2014;165(1):30–6.
Li B, Fan S, Yu F, Chen Y, Zhang S, Han F, Yan S, Wang L, Sun J. High-resolution mapping of QTL for fatty acid composition in soybean using specific-locus amplified fragment sequencing. Theor Appl Genet. 2017;130(7):1467–79.
Li X, Mei D, Liu Q, Fan J, Singh S, Green A, Zhou XR, Zhu LH. Down-regulation of crambe fatty acid desaturase and elongase in Arabidopsis and crambe resulted in significantly increased oleic acid content in seed oil. Plant Biotechnol J. 2016;14(1):323–31.
Liu F, Xia Y, Wu L, Fu D, Hayward A, Luo J, Yan X, Xiong X, Fu P, Wu G, Lu C. Enhanced seed oil content by overexpressing genes related to triacylglyceride synthesis. Gene. 2015;557(2):163–71.
Yu J, Zhang Z, Wei J, Ling Y, Xu W, Su Z. SFGD: a comprehensive platform for mining functional information from soybean transcriptome data and its use in identifying acyl-lipid metabolism pathways. BMC Genomics. 2014;15:271.
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–83.
Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, Corréa M, Da Silva C, Just J, Falentin C, Koh CS, Le CI, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao M, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier MC, Fan G, Renault V, Bayer PE, Golicz AA, Manoli S, Lee TH, Thi VH, Chalabi S, Hu Q, Fan C, Tollenaere R, Lu Y, Battail C, Shen J, Sidebottom CH, Wang X, Canaguier A, Chauveau A, Bérard A, Deniot G, Guan M, Liu Z, Sun F, Lim YP, Lyons E, Town CD, Bancroft I, Wang X, Meng J, Ma J, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury JM, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou Y, Hua W, Sharpe AG, Paterson AH, Guan C, Wincker P. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345(6199):950–3.
Wang L, Yu S, Tong C, Zhao Y, Liu Y, Song C, Zhang Y, Zhang X, Wang Y, Hua W, Li D, Li D, Li F, Yu J, Xu C, Han X, Huang S, Tai S, Wang J, Xu X, Li Y, Liu S, Varshney RK, Wang J, Zhang X. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014;15(2):R39.
Chen X, Li H, Pandey MK, Yang Q, Wang X, Garg V, Li H, Chi X, Doddamani D, Hong Y, Upadhyaya H, Guo H, Khan AW, Zhu F, Zhang X, Pan L, Pierce GJ, Zhou G, Krishnamohan KA, Chen M, Zhong N, Agarwal G, Li S, Chitikineni A, Zhang GQ, Sharma S, Chen N, Liu H, Janila P, Li S, Wang M, Wang T, Sun J, Li X, Li C, Wang M, Yu L, Wen S, Singh S, Yang Z, Zhao J, Zhang C, Yu Y, Bi J, Zhang X, Liu ZJ, Paterson AH, Wang S, Liang X, Varshney RK, Yu S. Draft genome of the peanut A-genome progenitor (Arachis duranensis) provides insights into geocarpy, oil biosynthesis, and allergens. Proc Natl Acad Sci U S A. 2016;113(24):6785–90.
Niu J, Chen Y, An J, Hou X, Cai J, Wang J, Zhang Z, Lin S. Integrated transcriptome sequencing and dynamic analysis reveal carbon source partitioning between terpenoid and oil accumulation in developing Lindera glauca fruits. Sci Rep. 2015;5:15017.
Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S, Zou C, Li Q, Yuan Y, Lu C, Wei H, Gou C, Zheng Z, Yin Y, Zhang X, Liu K, Wang B, Song C, Shi N, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Wang J, Yu S. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 2012;44(10):1098–103.
Zhang L, Wang SB, Li QG, Song J, Hao YQ, Zhou L, Zheng HQ, Dunwell JM, Zhang YM. An integrated bioinformatics analysis reveals divergent evolutionary pattern of oil biosynthesis in high- and low-oil plants. PLoS One. 2016;11(5):e0154882.
Ernst J, Bar-Joseph ZSTEM. A tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191.
Wan H, Cui Y, Ding Y, Mei J, Dong H, Zhang W, Wu S, Liang Y, Zhang C, Li J, Xiong Q, Qian W. Time-series analyses of transcriptomes and proteomes reveal molecular networks underlying oil accumulation in canola. Front Plant Sci. 2017;7:2007.
Jones SI, Vodkin LO. Using RNA-Seq to profile soybean seed development from fertilization to maturity. PLoS One. 2013;8(3):e59270.
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue):W316–22.
Ohlrogge J, Browse J. Lipid biosynthesis. Plant Cell. 1995;7(7):957–70.
Jin J, Zhang H, Kong L, Gao G, Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014;42(Database issue):D1182–7.
Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, Gao G. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45(D1):D1040–5.
Loic L, Jean V, Raymond C, Pierre G, Claude C. Phosphoenolpyruvate carboxylase: structure, regulation and evolution. Plant Sci. 1994;99(2):111–24.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server):W202–8.
Gargouri M, Park JJ, Holguin FO, Kim MJ, Wang H, Deshpande RR, Shachar-Hill Y, Hicks LM, Gang DR. Identification of regulatory network hubs that control lipid metabolism in Chlamydomonas reinhardtii. J Exp Bot. 2015;66(15):4551–66.
Lin H, Yu J, Pearce SP, Zhang D, Wilson ZA. RiceAntherNet: a gene co-expression network for identifying anther and pollen development genes. Plant J. 2017;92(6):1076–91.
Chung KO, Kim BY, Lee MH, Kim YR, Chung HY, Park JH, Moon JO. In-vitro and in-vivo anti- inflammatory effect of oxyresveratrol from Morus alba L. J Pharm Pharmacol. 2003;55(12):1695–700.
Cober ER, Voldenga HD. Developing high-protein, high-yield soybean populations and lines. Crop Sci. 2000;40(1):39–42.
Hu ZY, Hua W, Zhang L, Deng LB, Wang XF, Liu GH, Hao WJ, Wang HZ. Seed structure characteristics to form ultrahigh oil content in rapeseed. PLoS One. 2013;8(4):e62099.
Uhrig RG, O'Leary B, Spang HE, MacDonald JA, She YM, Plaxton WC. Coimmunopurification of phosphorylated bacterial- and plant-type phosphoenolpyruvate carboxylases with the plastidial pyruvate dehydrogenase complex from developing castor oil seeds. Plant Physiol. 2008;146(3):1346–57.
Gennidakis S, Rao S, Greenham K, Uhrig RG, O'Leary B, Snedden WA, Lu C, Plaxton WC. Bacterial- and plant-type phosphoenolpyruvate carboxylase polypeptides interact in the hetero-oligomeric Class-2 PEPC complex of developing castor oil seeds. Plant J. 2007;52(5):839–49.
Wang FL, Liu RH, Wu GT, Lang CX, Chen JQ, Shi CH. Specific down regulation of the bacterial-type PEPC gene by artificial microRNA improves salt tolerance in Arabidopsis. Plant Mol Biol Rep. 2012;30(5):1080–7.
O'Leary B, Rao SK, Kim J, Plaxton WC. Bacterial-type phosphoenolpyruvate carboxylase (PEPC) functions as a catalytic and regulatory subunit of the novel class-2 PEPC complex of vascular plants. J Biol Chem. 2009;284(37):24797–805.
Angelovici R, Lipka AE, Deason N, Gonzalez-Jorge S, Lin H, Cepela J, Buell R, Gore MA, Dellapenna D. Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell. 2013;25(12):4827–43.
Zhou M, Wang W, Karapetyan S, Mwimba M, Marqués J, Buchler NE, Dong X. Redox rhythm reinforces the circadian clock to gate immune response. Nature. 2015;523(7561):472–6.
Graf A, Schlereth A, Stitt M, Smith AM. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci U S A. 2010;107(20):9458–63.
Hsiao AS, Haslam RP, Michaelson LV, Liao P, Napier JA, Chye ML. Gene expression in plant lipid metabolism in Arabidopsis seedlings. PLoS One. 2014;9(9):e107372.
Yang J, Yang MF, Wang D, Chen F, Shen SH. JcDof1, a Dof transcription factor gene, is associated with the light-mediated circadian clock in Jatropha curcas. Physiol Plant. 2010;139(3):324–34.
Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G, Ohkura N, Azama T, Mesaki M, Yukimasa S, Kobayashi H, Iitaka C, Umehara T, Horikoshi M, Kudo T, Shimizu Y, Yano M, Monden M, Machida K, Matsuda J, Horie S, Todo T, Ishida N. Genome-wide expression analysis of mouse liver reveals clock-regulated circadian output genes. J Biol Chem. 2003;278(42):41519–27.
Daniel X, Sugano S, Tobin EM. CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci U S A. 2004;101(9):3292–7.
Madoka YK, Ken-Ichi T, Junya M, Ikuo N, Yukio N, Yukiko S. Chloroplast transformation with modified accD operon increases acetyl-CoA carboxylase and causes extension of leaf longevity and increase in seed yield in tobacco. Plant Cell Physiol. 2002;43(12):1518–25.
Konishi T, Shinohara K, Yamada K, Sasaki Y. Acetyl-CoA carboxylase in higher plants: most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol. 1996;37(2):117–22.
Davis MS, Solbiati J, Cronan JE. Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J Biol Chem. 2000;275(37):28593–8.
Li X, Ilarslan H, Brachova L, Qian HR, Li L, Che P, Wurtele ES, Nikolau BJ. Reverse-genetic analysis of the two biotin-containing subunit genes of the heteromeric acetyl-coenzyme a carboxylase in Arabidopsis indicates a unidirectional functional redundancy. Plant Physiol. 2011;155(1):293–314.
Schwender J, König C, Klapperstück M, Heinzel N, Munz E, Hebbelmann I, Hay JO, Denolf P, De Bodt S, Redestig H, Caestecker E, Jakob PM, Borisjuk L, Rolletschek H. Transcript abundance on its own cannot be used to infer fluxes in central metabolism. Front Plant Sci. 2014;5:668.
Gidda SK, Watt S, Collins-Silva J, Kilaru A, Arondel V, Yurchenko O, Horn PJ, James CN, Shintani D, Ohlrogge JB, Chapman KD, Mullen RT, Dyer JM. Lipid droplet-associated proteins (LDAPs) are involved in the compartmentalization of lipophilic compounds in plant cells. Plant Signal Behav. 2013;8(11):e27141.
Miquel M, Trigui G, d'Andréa S, Kelemen Z, Baud S, Berger A, Deruyffelaere C, Trubuil A, Lepiniec L, Dubreucq B. Specialization of oleosins in oil body dynamics during seed development in Arabidopsis seeds. Plant Physiol. 2014;164(4):1866–78.
Roos-Mattjus P, Sistonen L. The ubiquitin-proteasome pathway. Ann Med. 2004;36(4):285–95.
Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19(1):94–102.
Xu HM, Kong XD, Chen F, Huang JX, Lou XY, Zhao JY. Transcriptome analysis of Brassica napus pod using RNA-seq and identification of lipid-related candidate genes. BMC Genomics. 2015;16:858.
Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28(1):33–6.
Ritchie SW, Hanway JJ, Thompson HE, Benson GO. How a soybean plant develops. Iowa State University: Cooperative Extension Service; 1985. p. 1–20.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106.
Li L, Christian JS, David SR. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–89.
Van Dongen S. “Graph clustering by flow simulation.” Ph.D thesis. Utrecht: University of Utrecht; 2000.
Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: detection of (co-) orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124.
Sonnhammer EL, Östlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015;43(Database issue):D234–9.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57(1):289–300.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–21.
Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25(7):1253–6.
Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–5.
Yang Z. PAML. 4: a program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91.
Yang Z, Nielsen R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol. 1998;46(4):409–18.
Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–73.
Dai X. Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39(Web Server issue):W155–9.
Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(D1):D146–52.
Killcoyne S, Carter GW, Smith J, Boyle J. Cytoscape: a community-based framework for network modeling. Methods Mol Biol. 2009;563:219–39.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (31571268, 31871242), Huazhong Agricultural University Scientific & Technological Self-innovation Foundation (Program No. 2014RC020), and State Key Laboratory of Cotton Biology Open Fund (CB2017B01). Each of the funding bodies granted the funds based on a research proposal. They had no influence over the experimental design, data analysis or interpretation, or writing the manuscript.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Author information
Authors and Affiliations
Contributions
YZ conceived and supervised the study. ZZ carried out the experimental works and analyzed the data. ZZ, YZ and JMD wrote and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. 27,236 OGs of all the protein-coding genes in Arabidopsis thaliana, Glycine max, Brassica napus and Sesamum indicum. Table S2. Comparisons of sequence similarity-based protein families between Glycine max (gma) and Brassica napus (bna). Table S3. List of selected genes related to carbohydrate metabolism and lipid biosynthesis in Arabidopsis thaliana. Table S4. 230 candidate orthologous groups related to oil synthesis of seed for soybean, rapeseed and Arabidopsis. Table S5. Candidate orthologous groups (OGs) for the difference of seed oil content between rapeseed and soybean. Note: Genes with red color were negatively correlated with seed oil content and were obtained by cluster analysis of gene expression. The 44 OGs with black color differed in gene relative copy number between soybean and rapeseed. Table S6. Expressional contents and relative copy number of candidate genes associated with the difference of seed oil content in rapeseed and soybean. Note: The stages t1, t2, t3 and t4 were defined as R3, R4, R7 and R8 in soybean, and 2, 4, 6, and 8 weeks after pollination (WAP) in rapeseed, respectively. (XLSX 37341 kb)
Additional file 2:
Figure S1. The expression patterns for the genes of 230 gene families related to oil biosynthesis. The expression clustering analysis of 2048 soybean and rapeseed genes in the 230 gene families was performed using Short Time-series Expression Miner (STEM, http://www.cs.cmu.edu/~jernst/stem/) [56]. Here, t1 represents the seed oil initial synthesis stage; t2 to t3 represent the rapid accumulation period of seed oil biosynthesis; t4 represents the gradual decline stage after the seed oil accumulation content reaches the peak. In the end, all 2048 genes were clustered into 20 clusters. Figure S2. The expression profiles (A-D) of candidate genes related to oil biosynthesis. One down-regulated trend (profile 3) (A) and three up-regulated trends from t2 to t3 stages of seed oil biosynthesis (profile 13, 16 and 18, respectively) (B, C, D). Figure S3. Comparison of the expression patterns of the candidate genes between rapeseed and soybean. Note: t1-t4 and t1’-t4’ represent four seed development stages in rapeseed and soybean, respectively. PKp-α and PKp-β denote Alpha (α) and Beta (β) subunits of PK in plastid, respectively. ACCase contains homogeneous structure ACC2 and heterogeneous ACCase complex, which are composed of α-CT, β-CT, BC and BCCP. Figure S4. Comparison of the expression patterns of genes encoding enzymes PEPC, PK and ACCase. t1’, t2’, t3’ and t4’ represent R3, R4, R7 and R8 at soybean seed development stages, and t1, t2, t3 and t4 represent 2, 4, 6 and 8 weeks after pollination (WAP) at rapeseed seed development stages, respectively. Figure S5. Transcriptional regulation of key candidate genes for the difference of seed oil content between rapeseed and soybean. Figure S6. Evolutionary rate of each branch of PEPC gene family. ω0 = 0.340 represents the evolutionary rate when the evolutionary rate of each branch is assumed to be the same. (PDF 1291 kb)
Additional file 3:
Table S7. LRT results for selective pressure branch model (Model 0 vs two ratio model 2, df = 6). Table S8. LRT results for branch-site model (model A vs null model, df = 1). Table S9. Candidate genes for the differences of seed oil content between the two species and among cultivars in the same species. (PDF 93 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, Z., Dunwell, J.M. & Zhang, YM. An integrated omics analysis reveals molecular mechanisms that are associated with differences in seed oil content between Glycine max and Brassica napus. BMC Plant Biol 18, 328 (2018). https://doi.org/10.1186/s12870-018-1542-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-018-1542-8