Abstract
Background
Heat shock proteins 90 (HSP90s) are a highly conserved protein family of cellular chaperones widely found in plants; they play a fundamental role in response to biotic and abiotic stresses. The genome-wide analysis of HSP90 gene family has been completed for some species; however, it has been rarely reported for the tobacco HSP90 genes.
Results
In this study, we systematically conducted genome-wide identification and expression analysis of the tobacco HSP90 gene family, including gene structures, evolutionary relationships, chromosomal locations, conserved domains, and expression patterns. Twenty-one NtHSP90s were identified and classified into eleven categories (NtHSP90–1 to NtHSP90–11) based on phylogenetic analysis. The conserved structures and motifs of NtHSP90 proteins in the same subfamily were highly consistent. Most NtHSP90 proteins contained the ATPase domain, which was closely related to conserved motif 2. Motif 5 was a low complexity sequence and had the function of signal peptide. At least 6 pairs of NtHSP90 genes underwent gene duplication, which arose from segment duplication and tandem duplication events. Phylogenetic analysis showed that most species expanded according to their own species-specific approach during the evolution of HSP90s. Dynamic expression analysis indicated that some NtHSP90 genes may play fundamental roles in regulation of abiotic stress response. The expression of NtHSP90–4, NtHSP90–5, and NtHSP90–9 were up-regulated, while NtHSP90–6, and NtHSP90–7 were not induced by ABA, drought, salt, cold and heat stresses. Among the five treatments, NtHSP90s were most strongly induced by heat stress, and weakly activated by ABA treatment. There was a similar response pattern of NtHSP90s under osmotic stress, or extreme temperature stress.
Conclusions
This is the first genome-wide analysis of Hsp90 in N. tabacum. These results indicate that each NtHSP90 member fulfilled distinct functions in response to various abiotic stresses.
Similar content being viewed by others
Background
Plants are often affected by a variety of strenuous stresses during growth and development, including biotic and abiotic stresses, all of which are interrelated [1]. Moreover, the main abiotic stresses such as cold, drought, salinity, freezing, high light intensity, ozone (O3), and heat have a critical impact on the quality and yield of plants [2,3,4,5]. Recently, with global warming, heat stress has become one of the main abiotic stresses that affect the normal growth and development of plants all over the world [6,7,8].
Over the course of long-term evolution, plants form regulatory mechanisms which are resistant to adverse environmental conditions. When the plants are stimulated by heat or other factors, they produce highly conserved stress proteins called heat shock proteins (HSPs) [9,10,11]. Many types of HSPs have been identified in almost all organisms [12]. Heat shock proteins have been classified into HSP100/ClpB family, HSP90 family, HSP70/DnaK family, chaperonin (HSP60/GroEL) family, and small heat-shock proteins (sHSP) family based on their approximate molecular weights [3, 13,14,15].
Heat shock protein 90 family is a widespread class of molecular chaperones in eukaryotic cytoplasm, which is highly conserved [16,17,18]. For example, there are seven HSP90s in Arabidopsis, of which AtHSP90–1, AtHSP90–2, AtHSP90–3, and AtHSP90–4 are located in the cytoplasm, and AtHSP90–5, AtHSP90–6, and AtHSP90–7 are located in the chloroplast, mitochondria and endoplasmic reticulum, respectively [19, 20]. HSP90 is an ATP-regulated dimeric chaperone mainly consisting of three highly conserved domains: the C-terminal domain of about 25 kDa that binds to the substrate, the 35 kDa intermediate domain, and the 12 kDa N-terminal domain of the ATP-binding (NTD) [21,22,23]. HSP90s are part of the GHKL superfamily (HSP90, histidine kinase, MutL, and gyrases) of ATPases [24]. In HSP90s, the grooves that combine with ATP are often in a closed state [25], and the N-terminal ATPase activity is low [26]. When present in the cytoplasm of eukaryotic cells, HSP90s have a charged region between the middle domain and the N-terminal domain; the charged regions of different species have different lengths [27]. It is known that the function of any protein is determined by the formation and folding into a three-dimensional structure [28]. HSP90, as a class of chaperones, is mainly involved in the formation of the spatial structure of kinase substrate, DNA repair and substrate activation, initial stress signaling, the maintenance of the spatial structure of transcription factors, etc. [29,30,31,32,33]. Under stress or normal conditions, the HSP90 gene family has the function of preventing the aggregation of proteins and facilitating the refolding of inactive proteins [34], which together with other chaperones present in the organism forms a mechanism that assists in protein folding [3]. When plants are stressed, the expression of stressor HSP90 is up-regulated; it interacts with non-proteinaceous substances, and repairs the deformed protein [35].
Nine and seven HSP90 genes were found in Oryza sative [36] and Arabidopsis thaliana, respectively [19]. However, the identification of the tobacco HSP90 gene family has not yet been completed. Tobacco is an important economic crop and a typical model plant. Research on the tobacco HSP90 genes is of great significance for other plants [37]. The completion of genome-wide sequencing of tobacco provides the necessary information for data mining of HSP90 at the whole genome level [38]. In this study, we performed a genome-wide survey of Sol Genomics Network databases using HSP90 protein sequences from Arabidopsis. Bioinformatics methods were used to analyze gene structures, evolutionary relationships, chromosomal locations, and conserved domains of the tobacco HSP90 family in detail. In addition, we studied the expression patterns of the NtHSP90 genes under different abiotic stresses by qRT-PCR. The results are significant for the growth and development of tobacco and would provide a basis for further study of the biological functions of Hsp90 genes.
Results
Identification of the HSP90 gene family in Nicotiana tabacum
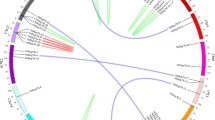
A local BLASTP search was used to identify HSP90 members in the tobacco genome using the Arabidopsis HSP90 protein sequence as a query sequence. We detected 21 predicted candidate HSP90 family proteins. In tobacco, the HSP90 genes were not randomly distributed on each chromosome; there were many gene clusters on the chromosome. Nine NtHSP90s were mapped onto 8 chromosomes and two NtHSP90s were located on chromosome 23. However, 12 NtHSP90s could not be conclusively localized to a chromosome (Fig. 1). In addition, there were at least 6 pairs of NtHSP90 genes that underwent gene duplication, which was possibly caused by segment duplication and tandem duplication events. Segment duplication resulted in many homologies of HSP90 genes between the chromosomes, which widened the HSP90 genome of tobacco. For example, Nitab4.5_0000152g0350 and Nitab4.5_0001622g0050 were the products of genomic segment replication.
The biophysical properties of coding HSP90s were calculated using the Expasy ProtParam tool. As shown in Table 1, the length of coding HSP90 sequences ranged from 594 to 2517 bp. The number of amino acids and biophysical properties of proteins encoded by different HSP90 genes were different, with amino acid number ranging from 197 to 838 on chromosomes. The molecular weight of different HSP90s varied greatly, and the fluctuation range was from 21,871.64 Da to 95,717.25 Da. The number of exons of the coding proteins ranged from 4 to 20. The isoelectric point (pI) of all HSP90s was acidic, indicating that the HSPs of tobacco were rich in acidic amino acids. Among them, the protein of Nitab4.5_0009637g0030 had the highest isoelectric point of 5.9430.
Phylogenetic analysis of HSP90 gene families
In order to further elucidate the evolutionary relationship of HSP90 gene families, an unrooted phylogenetic tree was constructed using the neighbor-joining method with the MEGA6.0 software, including seven HSP90s from Arabidopsis thaliana, eight from rice, six from tomato, and twenty-one from tobacco. The phylogenetic tree branch of Arabidopsis thaliana was consistent with the previous studies [19]; the seven members could be divided into five subfamilies. As shown in Fig. 2, the phylogenetic tree indicated that the HSP90s were clustered into ten clades (Clade 1 to 10). Notably, Clade 8 contained the HSP90s from four species (Arabidopsis thaliana, rice, tomato, and tobacco). The clade with the largest number of HSP90 genes was Clade 2, which were 2 from tobacco, 3 from rice, and 3 from Arabidopsis thaliana. In addition, 24 HSP90 genes were found to be homologous, accounting for 57.14% (24/42) of the total number of HSP90 genes. There were 12 pairs of paralogs within the species, one of which was from Arabidopsis thaliana (AT5G56000 and AT5G56010), two pairs from rice (Os09g30412 and Os09g30418, Os09g29840 and Os08g38086), and nine pairs from tobacco.
Exon-intron structure and phylogenetic analysis of the tobacco HSP90 gene family
Analysis of exon-intron structure can provide important insights into the evolution of gene families [39]. To analyze the exon-intron structure within the coding sequence in NtHSP90s, the genome and coding sequences of NtHSP90s were aligned using the Gene Structure Display Server (GSDS). A neighbor-joining phylogenetic tree was also constructed to explore whether the exon-intron distribution patterns correlate with the phylogenetic classification. The results showed that the HSP90 genes of tobacco could be clearly divided into eleven categories (NtHSP90–1 to NtHSP90–11; Fig. 3a). As shown in Fig. 3b, we found 11 different exon-intron distribution patterns, showing a high degree of similarity in the same branch. This conservation of exon-intron structure patterns in each class strongly supported the close evolutionary relationships of NtHSP90s in the tobacco HSP90 gene family; all NtHSP90s contained introns in the genomic sequences. The number of introns of the HSP90 genes varied greatly in tobacco. The HSP90 gene of NtHSP90–8 in the phylogenetic tree contained 19 introns, indicating that the gene was relatively complex. The HSP90 genes of NtHSP90–1, NtHSP90–2, NtHSP90–3, and NtHSP90–5 only contained 3 introns (Fig. 3b). The NtHSP90s usually varied in exon-intron distribution patterns and gene lengths in different clades.
Phylogenetic tree, exon-intron structure, and motif analysis of HSP90s. a Phylogenetic analysis of NtHSP90 proteins in tobacco. b Gene structure analysis of NtHSP90s. Exons and introns are indicated in black rectangles and black lines, respectively. c Conserved motifs analysis of NtHSP90 proteins using MEME tools. Conserved motifs are showed in different colored boxes
Multiple sequence alignment and C-terminal conserved motifs analysis
We analyzed all of the conserved motifs of NtHSP90s and identified the pattern of amino acid residues conservation in their domains. It was showed that NtHSP90s contained 10 conserved motifs, containing 21 to 50 amino acids (Table 2, Fig. 3c). Among them, motif 6 contained the least number of amino acids (21). Four motifs (motif 1, 2, 8, and 9) contained 50 amino acids. The motif 2 was found in all NtHSP90 members.
We performed a conserved motif analysis of NtHSP90s to obtain the pattern of amino acid residue conservation in NtHSP90 domains (Fig. 4). The results showed that the N-terminal domain was highly conserved, containing an ATPase site (red box). The conservative motif 2 was most widely distributed, which was closely related to the function of NtHSP90 N-terminal domain. Motif 2 constituted the ATPase domain of the NtHSP90 proteins, which functions as an ATP/ADP binding site with ATPase activity. Motif 5 was a low complexity sequence and had the function of signal peptide. Other motifs ensure the integrity of the NtHSP90 structures. Information of each motif, including motif logo and site number, was shown in Additional file 1: Figure S1.
Expression patterns analysis
Plants have formed a set of mechanisms for their own defense against stresses during long-term evolution. HSP90 genes are expressed as a response to abiotic stress [15]. To understand the expression patterns of NtHSP90s under abiotic stress, the transcript level of 11 NtHSP90s subclasses was analyzed by qRT-PCR. As shown in Fig. 5, different expression patterns of different NtHSP90s were observed under ABA, drought, salt, cold and heat stresses. The expression of NtHSP90–4, NtHSP90–5, and NtHSP90–9 were up-regulated, while NtHSP90–6, and NtHSP90–7 were not induced by the above-mentioned five treatments. ABA treatment induced the weakest stress response among the five treatments. Although ABA treatment induced the transcriptions of NtHSP90–4, NtHSP90–5, and NtHSP90–9 subclasses, the other 8 subclasses were not activated.
Expression patterns of NtHSP90s in response to ABA, PEG, NaCl, low and high temperature treatments. Transcript levels of NtHSP90s were analyzed by quantitative real-time PCR using L25 gene as an internal control. The unstressed expression level (0 h) was regarded as a standard because of its lowest expression. Values are the mean ± SE, n = 3
In response to drought (PEG treatment simulation) and NaCl treatments, there was a similar response pattern of NtHSP90s under osmotic stress. The transcriptions of NtHSP90–1, NtHSP90–2 and NtHSP90–3 were inhibited, whereas the transcript level of NtHSP90–4, NtHSP90–5, and NtHSP90–9 were increased. The remaining genes were not induced by drought and salt stress. The expression patterns of different members after PEG treatment were variable and the times of peak expression were also not consistent. For the NtHSP90–4, NtHSP90–5, and NtHSP90–9, the expression levels induced by NaCl treatment were stronger than those induced by PEG treatment.
There was a similar expression pattern of NtHSP90s in response to cold and heat stresses. Six subclasses of NtHSP90s (NtHSP90–4, NtHSP90–5, NtHSP90–8, NtHSP90–9, NtHSP90–10, and NtHSP90–11) were notably up-regulated under cold and heat stresses. Generally, high temperature treatment showed the strongest stress response among the five treatments. Under cold stress, NtHSP90–4, NtHSP90–5, and NtHSP90–9 showed high levels of transcription at 6–12 h; there was another peak at 48–72 h in expression levels of NtHSP90–8, NtHSP90–10, and NtHSP90–11. The expression patterns of six NtHSP90s under heat stress had unique expression profiles, responding with a single peak pattern showing high expression levels at 6–24 h.
Discussion
Understanding the response of plants to high temperature stress is very important for plant growth [40]. Therefore, it is necessary to identify genes that are involved in heat shock responses in plants. High temperature stress usually changes the expression of related genes in the organism. When the temperature is (5 °C) higher than the normal temperature, most of the normal protein synthesis and mRNA transcription in the organism is inhibited. However, at the same time, a class of highly conserved proteins called HSPs is quickly synthesized. HSPs were first identified in the salivary gland chromosomes of Drosophila larva [41]. Later, other studies found organisms produce a series of proteins of different sizes, known as HSPs, in response to increased temperatures [42]. In addition to high temperature stress, abiotic stresses such as drought, salinity, heavy metals, and ABA could also induce the production of HSPs in plants [43]. HSPs have been classified into HSP100, HSP90, HSP70, HSP60, and sHSP according to their approximate molecular weights [15]. Among them, HSP90 is an important and highly conserved HSP; it is a molecular chaperone widely found in eukaryotic cells [44]. HSP90 genes have been reported to be involved in kinase and transcription factor folding, stress signal transduction, and DNA repair [29, 45, 46]. They play an important role in maintaining and regulating the conformation and function of intracellular proteins. The HSP90 has been identified in many plant species. However, there is little information about HSP90 in tobacco. Here, we focused on the correlation analysis of the tobacco HSP90 genes. The comprehensive identification and characterization of the HSP90 gene family in tobacco was facilitated by the recent completion of tobacco genome sequencing. The identification and analysis of the tobacco HSP90 gene family will provide valuable insights into the genetic improvement of other plants.
In the present study, 21 HSP90 genes were isolated and identified from the common tobacco database using bioinformatics methods (Table 1). Nine genes were located on chromosomes (chromosomes 1, 2, 5, 8, 9, 12, 18, and 23). Compared with other comprehensive surveys of plant HSP90 gene family (7 HSP90 family genes have been identified in Arabidopsis, 9 in rice) [19, 36], the tobacco HSP90 gene family is the largest with 21 phylogenetic extension genes. It may be related to the fact that common tobacco is an allotetraploid plant [47], whose HSP90 genes have been replicated. These HSP90 family proteins play a key role in the physiological maintenance and environmental adaptability of tobacco, enabling it to survive in high temperature stress and other stressful environments. The different HSP90 family proteins have different biophysical properties, which further indicate that there is wide diversity among members, which will help to further study the function of HSP90 genes. In this study, the isoelectric points of tobacco HSP90 ranged from 3.8808 to 5.9430. Moreover, all tobacco HSP90 proteins were acidic, which was consistent with the results for Arabidopsis thaliana, tomato, and others [48]. The tobacco HSP90 genes were non-homogeneously distributed on chromosomes, mainly on both ends of the chromosome (Fig. 1), which was similar to the distribution of rice HSP90 genes [36]. Gene duplication is an important mechanism in the evolution of gene families [49]. There were at least 6 pairs of repeated genes identified in tobacco, indicating that gene replication may occur during the evolution of the tobacco HSP90 genes.
Phylogenetic analysis is usually used to obtain insight into the evolutionary relationships of species and to help identify orthologs between species and paralogs within species. In this study, an unrooted phylogenetic tree was constructed based on the full-length protein sequences of Arabidopsis, rice, tomato, and tobacco. According to phylogenetic analysis, HSP90s could be divided into ten clades (Fig. 2). The orthologous genes from Arabidopsis, rice, tomato, and tobacco were clustered in the same branch (Clade 8), indicating that NtHSP90s were primeval than the divergence of dicots and monocots. There were 12 pairs of paralogs within species, implying that most species expanded according to their own species-specific approach during the evolution of the HSP90 gene family. This finding was consistent with the findings for gene families of cereals such as rice [50, 51].
The structure of protein determines its function [52]. The amino acid sequence of HSP90 family proteins can provide phylogenetic relationship information based on its primary structure. We found that there were different numbers of introns (3 to 19 introns) in different NtHSP90 gene sequences. The number of introns is usually related to the sensitivity of gene transcription regulation. The lesser the number of introns, the stronger the plant’s ability to adapt to the diverse developmental processes and environmental stimuli [53]. The number of introns in NtHSP90s is the result of long-term evolution. According to the conserved motif analysis of the tobacco HSP90 genes, Nitab4.5_0001622g0050 and Nitab4.5_0003328g0120 contained fewer motifs, implying that it may have lost part of its sequence during evolution. Furthermore, there were 9 NtHSP90s that contained all 10 motifs, and their amino acid sequences were highly conserved. Motif 2 constituted the ATPase domain of the NtHSP90 proteins, which functions as an ATP/ADP binding site with ATPase activity [54]. The other nine motifs made up the tobacco HSP90 conserved domains, which play important roles in maintaining the complete ATPase domain activity [55].
Numerous studies have shown that the HSP90 genes are involved in response to abiotic stress [23, 56]. In the present study, we determined the dynamic expression levels of the NtHSP90 genes under ABA, drought, salt, cold and heat stresses. The results showed that the expression of NtHSP90–4, NtHSP90–5, and NtHSP90–9 were up-regulated, while NtHSP90–6, and NtHSP90–7 were not induced by the above-mentioned five treatments. Therefore, we speculate that NtHSP90–4, NtHSP90–5, and NtHSP90–9 are widely involved in the response to abiotic stresses, while NtHSP90–6, and NtHSP90–7 may not be involved in regulation of abiotic stress tolerance in tobacco. These results showed that individual NtHSP90 genes in the same clade may have distinct regulatory properties. High temperature treatment showed the strongest stress response among the different treatments, indicating that the NtHSP90 genes were more sensitive to high temperature stress response. The expression of NtHSP90s was induced by ABA, drought, salt, cold and heat stresses, which may reflect their potential roles in abiotic stress response.
Conclusions
In the present study, we systematically performed genome-wide identification and expression analysis of the tobacco HSP90 gene family, including gene structures, evolutionary relationships, chromosomal locations, conserved domains, and expression patterns. Twenty-one NtHSP90s were identified and classified into eleven categories. At least 6 pairs of NtHSP90 genes underwent gene duplication, which arose from segment duplication and tandem duplication events. Expression pattern analysis indicated that NtHSP90–4, NtHSP90–5, and NtHSP90–9 were induced by various abiotic stresses. NtHSP90s were strongly induced by heat stress, while weakly activated by ABA treatment. There was a similar response pattern of NtHSP90s under osmotic stress, or extreme temperature stress. The results provide a basis for further study of the biological functions of Hsp90 genes in response to abiotic stress.
Methods
Plant materials and stress treatments
The tobacco cultivar K326 (Nicotiana tabacum L., cv. Kentucky 326) was used for gene expression level related experiments. The tobacco seeds were sown in mixed soil (vermiculite:humus = 1:1) saturated with water in sieve-like plates. Seedlings were germinated and cultured in a greenhouse at 22 °C with a 16 h/8 h (light/dark) photoperiod for eight weeks. Then, they were assigned to a treatment group and separately stressed by exposure to a 50 μM ABA spray, a PEG 6000 solution (− 0.5 MPa), a 300 mM NaCl solution, a low temperature (4 °C), and a high temperature (42 °C), all of which were validated to cause a significant stress in pilot experiments. Untreated control plants were cultured normally. All the true leaves were sampled at 0, 1, 3, 6, 12, 24, 48, and 72 h after treatment. The main midribs were then removed. After collection, all samples were quickly frozen in liquid nitrogen and stored at − 80 °C for RNA isolation and analysis.
Identification of NtHSP90 genes
The Arabidopsis thaliana HSP90 proteome sequences were downloaded from the TAIR databases (http://www.arabidopsis.org/) [19]. The protein sequences of Arabidopsis thaliana HSP90 genes were used as query to perform BLASTP (E-value 1e-10) searched against N. tabacum genome sequences to obtain the final dataset of NtHSP90s (https://solgenomics.net/). Redundant sequences were removed. Then the Pfam (http://pfam.sanger.ac.uk/search) and SMART (http://smart.embl-heidelberg.de/) databases were used to confirm each predicted HSP90 protein [57, 58]. The sequences of rice HSP90s and tomato HSP90s were obtained from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) and the Solanaceae Genome Database (https://solgenomics.net/). The biophysical properties of coding HSP90s were calculated using the Expasy ProtParam tool (http://us.expasy.org/tools/protparam.html) [59].
Phylogenetic analysis
Multiple sequence alignment was carried out by using MUSCLE based on the sequences of Arabidopsis thaliana, rice, tomato, and tobacco [60]. The MEGA 6.0 software was used to construct an unrooted phylogenetic tree using the neighbor-joining method [61]. Support for the tree topology was evaluated by using a bootstrap analysis with 1000 replicates.
Gene structure analysis
A diagrammatic sketch of the HSP90 gene structure was constructed using the Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/) [62]; it was based on the alignment of the cDNAs with their corresponding genomic DNA sequences.
Multiple sequence alignment and motif analysis
Alignment of multiple HSP90 protein sequences from tobacco was performed using ClustalW (http://www.genome.jp/tools/clustalw/) [63]. The parameters were set to default values and the results of the alignment were visualized using the BoxShade program. The conserved motif of the full length of HSP90 family proteins was analyzed using the online MEME tool (Multiple Expectation Maximization for Motif Elicitation, http://meme-suite.org/tools/meme) [64]. The maximum motif search value was set at 10.
Chromosome distribution and synteny analysis
The distribution information of NtHSP90 gene family in the chromosome was obtained from the Sol Genomics Network (https://solgenomics.net/) database. For synteny analysis, synteny blocks containing HSP90 genes in the tobacco genome were identified using MCScanX program [65]. The chromosome distribution for each NtHSP90 and synteny relationship were displayed with circos (http://circos.ca/) [66].
Quantitative RT-PCR expression analysis of NtHSP90s
Total RNA was extracted from the samples using a modified CTAB method [67]. After removing genomic DNA contamination by DNase I (Fermentas, Waltham, MA, USA), 1 μg of total RNA was reverse-transcribed to cDNA using the PrimeScript™ RT Reagent Kit (Takara, Dalian, China). The PCR amplifications were performed using LightCycler® 480II (Roche Diagnostics, Indianapolis, IN, USA). For qRT-PCR, gene-specific primers were designed according to the cDNA sequences using Primer 6.0. Details of primers are shown in Additional file 2: Table S1. The transcription of tobacco ribosomal protein gene L25 (GenBank accession number L18908) was used as an internal reference gene. The qRT-PCR reactions were performed on the ABI 7900 HT Real-Time PCR System (Applied Biosystems) using the following thermal cycle: 95 °C for 5 min, followed by 40 cycles at 95 °C for 10 s, and 60 °C for 30 s. Three biological replicates were used for each gene. The relative expression level for each of NtHSP90s was calculated using the 2–ΔΔCT method [68].
Abbreviations
- ABA:
-
Abscisic acid
- HSP90:
-
Heat shock protein 90
- NtHSP90 :
-
Gene in Nicotiana tabacum
- PEG:
-
Polyethylene glycol
- pI:
-
Isoelectric point
- qRT-PCR:
-
Quantitative real-time PCR
References
Levitt J. Responses of plants to environmental stresses: academic press; 1980.
Chaves MM, Maroco JP, Pereira JS. Understanding plant responses to drought — from genes to the whole plant. Funct Plant Biol. 2003;30(3):239–64.
Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9(5):244–52.
Nakashima K, Yamaguchi-Shinozaki K. Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol Plant. 2006;126(1):62–71.
Bailey-Serres J, Voesenek LACJ. Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol. 2008;59(1):313–39.
Turner N, Kramer P: Adaptation of plants of water and high temperature stress: John Wiley & Sons; 1980.
Jagadish SV, Craufurd PQ, Wheeler TR. High temperature stress and spikelet fertility in rice (Oryza sativa L.). J Exp Bot. 2007;58(7):1627.
Moreno AA, Orellana A. The physiological role of the unfolded protein response in plants. Biol Res. 2011;44(1):75–80.
Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417(6889):618.
Robert: Evolution of heat shock proteins and immunity. 2002:449–464.
Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300(5622):1142.
Xiong L, Ishitani M: Stress signal transduction: components, pathways and network integration: springer Netherlands; 2006.
Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med. 2003;228(2):111–33.
Caplan AJ, Jackson S, Smith D. Hsp90 reaches new heights. Conference on the Hsp90 chaperone machine. Embo Reports. 2003;4(2):126.
Al-Whaibi MH. Plant heat-shock proteins: a mini review. Journal of King Saud University - Science. 2011;23(2):139–50.
Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH. Identification and structural characterization of the atp/adp-binding site in the hsp90 molecular chaperone. Cell. 1997;90(1):65–75.
Prasinos C, Krampis K, Samakovli D, Hatzopoulos P. Tight regulation of expression of two Arabidopsis cytosolic Hsp90 genes during embryo development. J Exp Bot. 2005;56(412):633–44.
Chen B, Zhong D, Monteiro A. Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genomics. 2006;7(1):156.
Krishna P, Gloor G. The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones. 2001;6(3):238–46.
Yamada K, Fukao Y, Hayashi M, Fukazawa M, Suzuki I, Nishimura M. Cytosolic Hsp90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J Biol Chem. 2007;282(52):37794–804.
Prodromou C. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90(1):65–75.
Terasawa K, Minami M, Minami Y. Constantly updated knowledge of hsp90. J Biochem. 2005;137(4):443.
Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75(75):271.
Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25(1):24.
Sung N, Lee J, Kim JH, Chang C, Joachimiak A, Lee S, Tsai FT. Mitochondrial Hsp90 is a ligand-activated molecular chaperone coupling ATP binding to dimer closure through a coiled-coil intermediate. Proc Natl Acad Sci U S A. 2016;113(11):201516167.
Raman S, Suguna K. Functional characterization of heat-shock protein 90 from Oryza sativa and crystal structure of its N-terminal domain. Acta Crystallogr. 2015;71(6):688–96.
Pearl LH, Prodromou C. Structure and in vivo function of Hsp90. Curr Opin Struct Biol. 2000;10(1):46–51.
Levitt M, Gerstein M, Huang E, Subbiah S, Tsai J. Protein folding: the endgame. Annu Rev Biochem. 1997;66(66):549–79.
Toft SEJaCQaDO. Hsp90: from structure to phenotype. Nature Structural and Molecular Biology. 2004;11:1152–5.
Shinozaki F, Minami M, Chiba T, Suzuki M, Yoshimatsu K, Ichikawa Y, Terasawa K, Emori Y, Matsumoto K, Kurosaki T. Depletion of hsp90β induces multiple defects in b cell receptor signaling. J Biol Chem. 2006;281(24):16361–9.
Zuehlke A, Johnson JL. Hsp90 and co-chaperones twist the functions of diverse client proteins. Biopolymers. 2010;93(3):211–7.
Rosa P, Paolo A, Alessandra DM. Hsp90: a new player in DNA repair? Biomolecules. 2015;5(4):2589–618.
Lachowiec J, Lemus T, Borenstein E, Queitsch C. Hsp90 promotes kinase evolution. Mol Biol Evol. 2015;32(1):91–9.
Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci. 2002;59(10):1640–8.
Mukhopadhyay I, Nazir A, Saxena DK, Chowdhuri DK. Heat shock response: hsp70 in environmental monitoring. J Biochem Mol Toxicol. 2003;17(5):249–54.
Hu W, Hu G, Han B. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 2009;176(4):583–90.
Brisson N, Ruget F, Gate P, Lorgeou J, Nicoullaud B, Tayot X, Plenet D, Jeuffroy MH, Bouthier A, Ripoche D: STICS [Simulateur multidisciplinaire pour les Cultures Standards]: a generic model for simulating crops and their water and nitrogen balances. 2. Model validation for wheat and maize. Agronomie 2002.
Sierro N, Battey JND, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov NV. The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun. 2014;5(5):3833.
Mach J. Alternative splicing produces a JAZ protein that is not broken down in response to jasmonic acid. Plant Cell. 2009;21(1):14.
Braun JW, Khan AA. Alleviation of salinity and high temperature stress by plant growth regulators permeated into lettuce seeds via acetone. Journal American Society for Horticultural Science. 1976;101(6):716–21.
Ritossa F. A new puffing pattern induced by temperature shock and DNP in drosophila. Experientia. 1962;18(12):571–3.
Tissières A, Mitchell HK, Tracy UM. Protein synthesis in salivary glands of drosophila melanogaster: relation to chromosome puffs. J Mol Biol. 1974;84(3):389.
Vierling E. The roles of heat shock proteins in plants. Annurevplant Physiolplant Molbiol. 1991;42(1):579–620.
Bose S, Weikl T, Bügl H, Buchner J. Chaperone function of Hsp90-associated proteins. Science. 1996;274(5293):1715.
Wegele H, Müller L, Buchner J. Hsp70 and Hsp90--a relay team for protein folding. Revphysiolbiochempharmacol. 2004;151(1):1–44.
Shinozaki F, Minami M, Chiba T, Suzuki M, Yoshimatsu K, Ichikawa Y, Terasawa K, Emori Y, Matsumoto K, Kurosaki T. Depletion of hsp90beta induces multiple defects in B cell receptor signaling. J Biol Chem. 2006;281(24):16361–9.
Lim KY, Matyasek R, Kovarik A, Leitch AR. Genome evolution in allotetraploid Nicotiana. Biol J Linn Soc. 2015;82(4):599–606.
Liu Y, Wan H, Yang Y, Wei Y, Li Z, Ye Q, Wang R, Ruan M, Yao Z, Zhou G: Genome-wide identification and analysis of heat shock protein 90 in tomato. Yi chuan =Hereditas 2014, 36(10):1043–1052.
Yang Z, Zhou Y, Wang X, Gu S, Yu J, Liang G, Yan C, Xu C. Genomewide comparative phylogenetic and molecular evolutionary analysis of tubby-like protein family in Arabidopsis, rice, and poplar. Genomics. 2008;92(4):246–53.
Bai J, Pennill LA, Ning J, Lee SW, Ramalingam J, Webb CA, Zhao B, Sun Q, Nelson JC, Leach JE. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 2002;12(12):1871.
Jain M, Tyagi AK, Khurana JP. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics. 2006;88(3):360–71.
Skolnick J, Fetrow JS. From genes to protein structure and function: novel applications of computational approaches in the genomic era. Trends Biotechnol. 2000;18(1):34–9.
Jin Z, Chandrasekaran U, Liu A. Genome-wide analysis of the Dof transcription factors in castor bean ( Ricinus communis L.). Genes and Genomics. 2014;36(4):527–37.
Young JC, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 2001;154(2):267–73.
Philippe M, Chrisostomos P, Bin H, Vaughan Mark S. Structural and functional analysis of the middle segment of Hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11(3):647–58.
Panaretou B, Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 2014;17(16):4829–36.
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296.
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangradorvegas A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279.
Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, Castro ED, Flegel V, Fortier A, Gasteiger E, Ioannidis V. ExPASy: SIB bioinformatics resource portal. Nucleic acids research. 2012;40(Web Server issue):597–603.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9.
Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic acids research. 2006;34(Web Server issue):W609.
Rédei GP: CLUSTAL W (improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice): springer Netherlands; 2008.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME Suite: tools for motif discovery and searching. Nucleic acids research. 2009;37(Web Server issue):202–8.
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee T, Jin H, Marler B, Guo H. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49.
Krzywinski M, Schein JI. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–45.
Wan SB, Tian L, Tian RR, Pan QH, Zhan JC, Wen PF, Chen JY, Zhang P, Wang W, Huang WD. Involvement of phospholipase D in the low temperature acclimation-induced thermotolerance in grape berry. Plant Physiol Biochem. 2009;47(6):504–10.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–8.
Acknowledgments
We appreciate the reviewers and editors for the patience to the work.
Funding
This work was supported by National Natural Science Foundation of China (NSFC31301837) and China National Tobacco Corp., Chongqing Division (NY20150601070011). They provided support for data collection and manuscript writing.
Availability of data and materials
The data sets supporting the results of this article are included within the article and its additional file.
Author information
Authors and Affiliations
Contributions
ZPS, FLP and HYZ conceived and designed the experiments. CY, HLJ and NJL performed the experiments and participated to the data analysis. XCL performed the qRT-PCR experiments. HFJ and FH revised the manuscript. All authors have read and approved the manuscript, and ensure that this is the case.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The tobacco cultivar K326 (Nicotiana tabacum L., cv. Kentucky 326) was used in the present study. The K326 originated from a cross of two breeding lines which obtained from the cross breeding of Coker 139, Coker 319 and McNair 30, NC 95, respectively, and released in 1982 by Novartis Seeds, Inc.. The K326 seeds used in this study were purchased from the Tobacco Research Institute of the Chinese Academy of Agricultural Sciences in Qingdao, China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. Motif analysis of the NtHSP90 proteins. The 10 motifs were analyzed using the MEME online tool. Different letters represent the abbreviation of various amino acids. The higher the letter height, the stronger the conservatism of the amino acid at that position. (PPTX 417 kb)
Additional file 2:
Table S1. Specific primers of NtHSP90 in qRT-PCR. (DOCX 16 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Song, Z., Pan, F., Yang, C. et al. Genome-wide identification and expression analysis of HSP90 gene family in Nicotiana tabacum. BMC Genet 20, 35 (2019). https://doi.org/10.1186/s12863-019-0738-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12863-019-0738-8