Abstract
Background
Whole genome duplications (WGDs) have been proposed to have made a significant impact on vertebrate evolution. Two rounds of WGD (1R and 2R) occurred in the common ancestor of Gnathostomata and Cyclostomata, followed by the third-round WGD (3R) in a common ancestor of all modern teleosts. The 3R-derived paralogs are good models for understanding the evolution of genes after WGD, which have the potential to facilitate phenotypic diversification. However, the recent studies of 3R-derived paralogs tend to be based on in silico analyses. Here we analyzed the paralogs encoding teleost olfactory marker protein (OMP), which was shown to be specifically expressed in mature olfactory sensory neurons and is expected to be involved in olfactory transduction.
Results
Our genome database search identified two OMPs (OMP1 and OMP2) in teleosts, whereas only one was present in other vertebrates. Phylogenetic and synteny analyses suggested that OMP1 and 2 were derived from 3R. Both OMPs showed distinct expression patterns in zebrafish; OMP1 was expressed in the deep layer of the olfactory epithelium (OE), which is consistent with previous studies of mice and zebrafish, whereas OMP2 was sporadically expressed in the superficial layer. Interestingly, OMP2 was expressed in a very restricted region of the retina as well as in the OE. In addition, the analysis of transcriptome data of spotted gar, a non-teleost fish, revealed that single OMP gene was expressed in the eyes.
Conclusion
We found distinct expression patterns of zebrafish OMP1 and 2 at the tissue and cellular level. These differences in expression patterns may be explained by subfunctionalization as the model of molecular evolution. Namely, single OMP gene was speculated to be originally expressed in the OE and the eyes in the common ancestor of all Osteichthyes (bony fish including tetrapods). Then, two OMP gene paralogs derived from 3R-WGD reduced and specialized the expression patterns. This study provides a good example for analyzing a functional subdivision of the teleost OE and eyes as revealed by 3R-derived paralogs of OMPs.
Similar content being viewed by others
Background
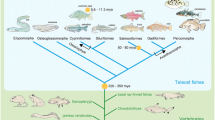
Gene duplication is one of the major driving forces of evolution [1–3]. In particular, whole genome duplication (WGD) has been thought to be an important factor in the evolution of vertebrates [1]. It has been proposed that at least two rounds of WGDs occurred during the evolution of vertebrates [1, 4, 5] (Fig. 1). Recent genome studies of amphioxus [6] and lamprey [7], support this hypothesis. The first-and second-round WGD (1R and 2R, respectively) are suggested to have occurred in the common ancestor of Gnathostomata and Cyclostomata [7, 8]. Thus, almost all modern vertebrates are believed to have undergone WGDs at least twice [1]. The third-round WGD (3R), in contrast, occurred in the common ancestor of teleosts (ray-finned fish excluding basal groups belong to polypteriforms, acipenseriforms, lepisosteids, and Amia) [8–12]. This is represented by the copy number of genes, two in teleosts, one in mammals [12, 13]. The Hox cluster is the most well-known example in this regard [9–11]. There are seven Hox clusters in teleost genomes, whereas four clusters are present in mammalian, coelacanth, and shark genomes [11]. In addition, slightly differentiated expression patterns are observed for teleost Hox paralogs derived from 3R [14–17]. Thus, teleost-specific duplicated genes seem to be on a path to functional differentiation, namely, 3R occurred neither too recently nor too early to differentiate the function of paralogs. 3R-derived paralogs could be good examples for evaluating the critical timing of functional differentiation.
In the case of gene duplication, it is traditionally expected that one of the duplicated genes becomes free from selective pressure and accumulates mutations in the protein coding and/or cis regulatory regions that led to the loss of its functions (pseudogenization) or gain of new functions (neofunctionalization) [2, 3]. Alternatively, subfunctionalization is observed especially as a result of WGD. In subfunctionalization, both paralogs are functional, but each paralog undergoes a complementary reduction and specialization in its expression pattern because of the mutation of its cis regulatory regions [3, 18, 19]. The subfunctionalized paralogs are also expected to gain new function over evolutionary time [20]. Most of the different expression patterns between teleost-specific paralogs that have been shown by previous studies [14–17, 21] could be explained by subfunctionalization. However, in these studies, the different expression patterns were usually discussed based only on in silico studies. In cases where analyses were carried out in vivo, many were performed only at the whole-mount tissue level. Very few comparative expression analyses have been carried out at the cellular level, which is indispensable for the investigation of subfunctionalization.
Olfactory marker protein (OMP) was first isolated from mouse olfactory bulb in the 1970s [22]. OMP is a small protein (~20 kDa) that consists of ~160 amino acids and is specifically expressed in olfactory sensory neurons (OSNs), which are distributed in the main olfactory epithelium in various vertebrates [22–24]. Although OMP is used as a specific marker of mature OSNs in vertebrates [25–28], its function has not yet been fully elucidated. OMP-knockout (KO) mice have reduced physiological activity and behavioral responses with respect to sensing odorants as compared with wild-type mice [29, 30]. In addition, axons of OSNs from OMP-KO mice project abnormally [31]. Recent studies have suggested that OMP is a critical factor for the functional maturation of OSNs [32] and is likely to be involved in Ca2+ clearance in OSNs [33]. Namely, the phenotypes observed in OMP-KO mice mentioned above might be caused by a decline in the clearance of Ca2+ in these OSNs.
OMP had been believed to be a highly conserved single-copy intronless gene among all vertebrates [24, 27, 34, 35]. More recently, it was shown that African clawed frog (Xenopus laevis) and some teleosts have two OMPs [36–38]. In African clawed frog, the expression patterns of the two OMPs are notably, albeit not completely, distinct in the lateral diverticulum and medial diverticulum, in the nasal cavity [36]. These expression patterns are suggestive of subfunctionalization. Although the expression of each of the two OMPs was analyzed in medaka (Oryzias latipes) [37] and salmon (Oncorhynchus nerka) [38], detailed expression patterns were not assessed. In this study, we found with a bioinformatic analysis that teleosts generally possess two OMPs in their genomes. Our phylogenetic analyses revealed that two OMPs are derived from 3R. Until now, expression pattern of OMP has been investigated in many vertebrates. Accordingly, we expected that detail verification of the expression patterns of OMPs could be a good example to understand the fate of 3R-derived paralogs. We here shows the detailed expression patterns of two OMPs at the tissue and the cellular level in zebrafish (Danio rerio).
Results
Two OMPs derived from the third-round whole genome duplication in teleosts
To identify OMPs in teleost genomes, we searched genome databases of zebrafish, stickleback (Gasterosteus aculeatus), fugu (Takifugu rubripes), medaka, platyfish (Xiphophorus maculatus), and tilapia (Oreochromis niloticus) by using known OMP sequences as queries, and obtained two significant hits from each species (see Methods for Data mining). Although zebrafish was believed to have a single copy of OMP [27], we isolated two OMPs from the genomes of all teleost, including zebrafish. We named the already-known zebrafish OMP as OMP1 and its paralog as OMP2. Furthermore, Ensembl gene prediction suggested that OMP2 consists of two exons, although OMP is known as an intronless gene [24, 36]. We searched OMP2 sequences from the EST database and confirmed that the predictions are consistent with the mRNA sequences in zebrafish, stickleback, and medaka. In other teleost species, we estimated the OMP2 gene structure with GeneWise. We also searched other vertebrate genome databases and isolated OMP orthologs. Then, we aligned the amino acid sequences of these OMP homologs (Fig. 2). Amino acids sequences are conserved among teleost OMP1 and OMP2 and tetrapod OMP. In particular, the Eph2B-receptor-like loop domain, a potentially key region for OMP function as a molecular switch [35], is highly conserved. Thus, the fundamental structure and physiological function of OMP2 are expected to be similar to those of OMP1 or tetrapod OMP.
Sequence analysis of OMP. Alignment of amino acid sequences was constructed by ClustalW2. Colors indicate OMP subgroups: blue, teleost OMP1; red, teleost OMP2; purple, gar OMP; black, tetrapod OMP. Hyphens indicate gaps. Light characters indicate conserved amino acids among homologs. Vertical dashed line is the junction of OMP2 exon 1 and exon 2. Secondary structure is based on Smith et al. [35]. OMP has two α-helical regions and eight β-pleated sheets, as shown in the structure below the sequences
Based on the genome search, we found that only one OMP exists in gar (Lepisosteus oculatus), which diverged from the teleosts before the occurrence of 3R [12]. These data suggest that the two OMPs in teleosts were derived from 3R. To investigate this possibility, we performed phylogenetic analysis that included gar and tetrapods. We constructed a maximum likelihood phylogenetic tree using amino acids sequences from exon 2 of OMP2 and the homologous regions of OMP1 and tetrapod OMP (Fig. 3). Teleost OMP homologs formed a monophyletic group with a bootstrap value of 99 % that consisted of the OMP1 clade (a bootstrap value of 66 %) and OMP2 clade (a bootstrap value of 99 %). These data strongly suggest that OMP was duplicated in a common ancestor of teleosts, after the divergence of gar. Thus, we suggest that teleost-specific OMP duplication was derived from 3R. It should be noted that each of the salmon OMPs and xenopus OMPs formed a monophyletic group, suggesting that duplication of salmon OMPs and xenopus OMPs was caused by lineage-specific WGDs that occurred independently in those two lineages.
We next analyzed the synteny of OMP loci (Fig. 4) and found that OMP is located within the intron of another gene, Calpain5 (CAPN5). Interestingly, teleost CAPN5, together with OMP, was also duplicated. According to the ZFIN [39], CAPN5a are encoded on chromosome 18 and CAPN5b are encoded on chromosome 21. Consequently, OMP1 is linked to CAPN5b and OMP2 is linked to CAPN5a. Given that both OMP1 and 2 are located within intron2 of CAPN5b and a, respectively, it is highly unlikely that OMP duplication was caused by retrotransposition. The genomic structures around OMPs (~40 kb) are well conserved between paralogs and also among species except for the coding direction of OMP. The results of the synteny analysis support our expectation that the two OMPs are derived from 3R.
Synteny analysis of OMP loci. Exon map was drawn based on Ensembl annotations. Boxes indicate exons: those above the line indicate forward strand-coded; those under the lines indicate reverse strand-coded. Colors indicate OMP or CAPN5 subgroups: blue, teleost OMP1; cyan, teleost CAPN5a; red, teleost OMP2; pink, teleost CAPN5b; purple, gar OMP; light purple, gar CAPN5; black, tetrapod OMP; gray, tetrapod CAPN5
OMP2 expression in the retina
Three typical fates of duplicated genes are known: pseudogenization, neofunctionalization, and subfunctionalization [3, 18, 19]. There is another fate of duplicated genes, in which gene duplication simply increases the amount of products as represented by the ribosomal DNA genes [40]. However, this is an extreme case in that more than hundred copies exist in the genome. Accodingly, we focus on the possibilities of neofunctionalization and subfunctionalization, which are generally accompanied by differentiated expression patterns that can be assessed by in situ hybridization. We thus examined the expression patterns of OMP1 and OMP2 (Fig. 5). At first, we investigated the expression of zebrafish OMP1 and OMP2 by RT-PCR with total RNA extracted from each organ as template (Fig. 5a). OMP1 was specifically expressed in the OE, whereas OMP2 was expressed in the eyes as well as the OE. The expression of OMP2 in the retina is quite interesting because OMP was believed to be specifically expressed in the olfactory organ [24, 27, 36]. To examine whether the expression of OMP2 in the retina is a common phenomenon among other teleost species, we searched the teleost EST database and found that the expression of OMP2 is detected in the eyes or the retina of stickleback and tilapia (Table 1), showing that OMP2 is expressed not only in zebrafish eyes but also in the eyes of some other teleosts. We also examined the expression of OMP in spotted gar, of which transcriptome data from the eyes is available. Interestingly, OMP was shown to be apparently expressed in the eyes (Fig. 5b). Next, we performed fluorescence in situ hybridization with antisense riboprobe (Fig. 5c) or sense riboprobe (Fig. 5d) to OMP2 to examine expression patterns in detail using cryosectioned eye tissues. Surprisingly, OMP2-positive signals were detected specifically in the outermost part (Fig. 5c, arrow) of the inner nuclear layer, where retinal horizontal cells are distributed [41].
Expression patterns of OMP1 and 2 at the tissue level. a RT-PCR analysis of OMP1, OMP2, and GAPDH (positive control) in adult zebrafish. DNA templates were as follows: B, brain; OE, olfactory epithelium; E, eye; G, gill; S, skin; T, testis; O, ovary; Gen, genomic DNA. RT–, the cDNA synthesis was performed without reverse transcriptase as a negative control for RT-PCR of OMP1. Note: When genomic DNA was used as a tmplates, PCR using OMP2 or GAPDH primers did not amplified fragments because of the presence of introns (over 2 kb) in the corresponding sequences in the genomic DNA. b Expression of OMP genes in eyes of gar and zebrafish. Bars indicate FPKM ratio of each OMP to Gαt1, which is coupling with rhodopsins. The number of spotted gar indicate technical replicate. c, d Fluorescence in situ hybridization analysis for OMP2 using DIG-labeled antisense riboprobes (c) or sense riboprobes (d) in transverse sections of adult zebrafish eyes, which were counterstained with DAPI. GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer. Arrow indicates OMP2-expressing zone. Scale bar, 50 μm
Divergence of expression patterns between OMP1 and OMP2 in the OE
RT-PCR analysis (Fig. 5a) showed that both OMP1 and 2 are expressed in the OE. To investigate these expression patterns at the cellular level, we performed two-color fluorescence in situ hybridization using separately labeled antisense riboprobes (Fig. 6a-c). Zebrafish OMP (synonym, OMP1) was known as a molecular marker for the ciliated OSNs, and was broadly expressed in the deep layer of the olfactory placode (Fig. 6a) as Sato et al. reported previously [28]. In contrast, OMP2 was sparsely expressed in the superficial layer (Fig. 6b). Merged images show highly exclusive expression of OMP1 and 2 (Fig. 6c). There were fewer OMP2-expressing cells than OMP1-expressing cells. These results showed that OMP1 and 2 are mainly expressed in the deep and superficial layer, respectively, and have distinct expression patterns at the cellular level in the OE. In addition, we noticed that a few cells in the superficial layer expressed both OMP1 and 2 (Fig. 6a-c, arrowheads). Thus, OMP1 was also expressed infrequently in the superficial layer and the OMP1-expressing cells in the superficial layer coexpressed OMP2. Previously, Sato et al. [28] also reported that zebrafish TRPC2 is a marker for the microvillous OSNs, and was expressed in the superficial layer of the OE. We therefore examined the expression of OMP1 and TRPC2 (Additional file 1: Figure S1). As expected, OMP1 was expressed in the deep layer (Additional file 1: Figure S1A), whereas TRPC2 was in the superficial layer (Additional file 1: Figure S1B). Merged images showed that these genes were not coexpressed (Additional file 1: Figure S1C). We also confirmed that OMP1 was sparsely expressed in the superficial layer (Additional file 1: Figures S1A-C, arrows). Next, we analyzed the expression of TRPC2 and OMP2 (Fig. 6d-f). Merged images showed that expression of these did not overlap (Fig. 6f), indicating that OMP2 was not coexpressed with TRPC2, although both genes were expressed in the superficial layer. The distinctive expression of OMP2 might suggest that OMP2-expressing cells are not OSNs. Thus, we examined the expression of NCAM, a neural marker, and OMP2 (Fig. 6g-i). Merged images showed that OMP2 was coexpressed with NCAM (Fig. 6g-i, arrowheads). The results strongly suggest that OMP2-expressing cells are actually OSNs.
Expression patterns of OMP1 and 2 at the cellular level in the OE. Two-color fluorescence in situ hybridization analysis using DIG- or fluorescein-labeled antisense riboprobes in horizontal sections of the adult zebrafish OE. a, d, g Fluorescent images of Alexa 594 derived from DIG-labeled riboprobes. b, e, h Fluorescent images of Alexa 488 derived from fluorescein-labeled riboprobes. c, f, i Merged images of (a) and (b), (d) and (e), and (g) and (h), respectively. Yellow two-headed arrows and cyan two-headed arrows indicate the superficial layer and the deep layer, respectively. Dashed lines indicate the outlines of the epithelium. White arrowheads indicate cells that coexpress OMP2 and another gene. Scale bar, 20 μm
G-protein coexpressed with OMP1 and OMP2
OMP2 is expected to be expressed in unidentified OSNs. We are interested in which olfactory receptor genes are coexpressed. Now, four types of olfactory receptor genes are known: odorant receptors (ORs) [42], trace amine-associated receptors (TAARs) [43], vomeronasal type 1 receptors (V1Rs) and vomeronasal type 2 receptors (V2Rs) [44–46]. However, it is technically hard to examine the coexpression of OMP2 with receptor genes, because the copy number of them are very large. We thus focused on G-protein α-subunits (Gα) genes. It was simply believed that Gαolf is coupled with both ORs [42] and TAARs [43], Gαo is coupled with V2Rs [45, 46], and Gαi2 is coupled with V1Rs [45, 46]. Oka et al. [47] have shown that some Gα families are also duplicated in teleosts and are expressed in the OE, namely Gαolf2, Gαo1, Gαo2, and Gαi1b (synonym, gnal, gnao1a, gnao1b, and gnaia, respectively) are expressed in the sensory area of the zebrafish OE. We performed the confirmatory analyses for the expression of the above genes by fluorescent in situ hybridization. We also exploratory chose four additional Gα genes, Gαi1a, Gαi2a, Gαi2b, and Gαq (synonym, gnai1, gnai2b, gnai2a, and gnaq, respectively), which seem to be well expressed in OE in RT-PCR [47] for the in situ hybridization analyses. We were able to detect clear signals for only Gαolf2, Gαo2, and Gαi1b; for the five other genes were not detected (data not shown), probably because the expressions levels of these genes were too low and/or the number of cells expressing these genes were too small. First, we examined the expression of Gαolf2 and Gαo2 (Additional file 2: Figure S2). Gαolf2 was mainly expressed in the deep layer (Additional file 2: Figure S2A), whereas Gαo2 was expressed in the superficial layer (Additional file 2: Figure S2B). Merged images showed that Gαolf2 and Gαo2 were not coexpressed (Additional file 2: Figure S2C). It should be noted that a few Gαolf2-expressing cells were in the superficial layer (Additional file 2: Figures S2A-C arrows). The expression patterns of Gαolf2 and Gαo2 are similar to those of OMP1 and TRPC2 (Additional file 1: Figure S1), respectively. We also confirmed that OMP1-positive signals frequently overlapped with Gαolf2-positive signals (Additional file 3: Figure S3). Second, we examined the expression of OMP2 and three Gα genes (Fig. 7). Interestingly, OMP2-positive signals overlapped with the Gαolf2-positive signals in the superficial layer (Fig. 7a-c, arrowheads), indicating that OMP2 was expressed in the Gαolf2-expressing cells whose cell bodies were situated in the superficial layer. In contrast, OMP2 was not expressed in Gαo2-expressing cells, although both genes were expressed in the superficial layer (Fig. 7d-f). OMP2 was not expressed in the Gαi1b-expressing cells, which were spottily and sparsely situated in the OE (Fig. 7g-i). These results strongly suggest that OMP2-expressing cells coexpress Gαolf2. In addition, we examined the coexpression of OMP2 with Ora genes, which are similar to V1Rs and retained only 6 copies in teleosts [48, 49], but none of them was coexpressed with OMP2 (Additional file 4: Figure S4).
Coexpression of OMP2 and Gα families. Two-color fluorescence in situ hybridization analysis using DIG- or fluorescein-labeled antisense riboprobes in horizontal sections of the adult zebrafish OE. a, d, g Fluorescent images of Alexa 594 derived from DIG-labeled riboprobes. b, e, h Fluorescent images of Alexa 488 derived from fluorescein-labeled riboprobes. c, f, i Merged images of (a) and (b), (d) and (e), and (g) and (h), respectively. Yellow two-headed arrows and cyan two-headed arrows indicate the superficial layer and the deep layer, respectively. Dashed lines indicate the outlines of the epithelium. White arrowheads indicate cells that coexpress OMP2 and another gene. Scale bar, 20 μm
Discussion
Novel insight into the function of OMP in the visual system
Here we report that the two OMPs in teleosts are derived from 3R and have functionally diverged over the ensuing years. We showed the obviously non-canonical expression patterns of OMP2 in the eyes of broad teleost species (Fig. 5, Table 1). OMP2 appeared to be expressed in retinal horizontal cells in zebrafish (Fig. 5c). The expression of OMP2 in the eyes is very interesting because OMP has been believed to be an olfactory organ-specific protein [24, 27, 36]. Unexpected finding of OMP2 expression in the retina prompted us to investigate the OMP of non-teleost fish to understand the ancestral state. We showed that relative expression level of gar OMP was as high as that of zebrafish OMP2 in the eye (Fig. 5b). This data underlies that the OMP has been already expressed in the eyes of the common ancestor of bony fish. A recent study reported that OMP is expressed in mouse cornea and proposed that OMP might be involved in the developmental process of corneal epithelial cells [50]. We searched EST database of mouse and xenopus, only to find no OMP sequence from the eye or the retina (data not shown). We also analyzed OMP2 expression in zebrafish cornea by in situ hybridization and detected no OMP2-positive signals in the cornea (data not shown). We thus believe that OMP2 is not expressed in cornea but expressed in retina of zebrafish. Although there is slight discrepancy between mice and zebrafish in that the OMP expression is detected in cornea of mice whereas OMP2 in retina of zebrafish, the expression of OMP gene in the visual system is expected to be an ancestral state (Fig. 8, Additional file 5: Figure S5). Based on the above lines of evidence, we propose that the expression of OMP2 in visual system of teleosts could be explained by subfunctionalization (Fig. 8).
OMP is colocalized with Na+/Ca2+ exchanger 1 (NCX1) and is involved in the mechanism of Ca2+ clearance in mouse OSNs [33]. The NCX1 ortholog is duplicated in zebrafish, and one of these, NCX1b, is expressed in zebrafish eyes as well as in other neural tissues [51, 52]. Thus it is possible that OMP2 and NCX1b are colocalized and are both involved in the regulation of cations in teleost retinal horizontal cells. To further understand the mechanism underlying subfunctionalization in eyes caused by 3R, it is worth examining the gene expressions of OMP and NCX1 of non-teleost fish as well as mice.
Characterization of OMP1- and OMP2-expressing OSNs
Teleost OE contains three types of OSNs: ciliated, microvillous, and crypt OSNs [53–55]. Generally, the cell bodies of the ciliated OSNs are situated in the deep layer of the OE, whereas those of microvillous OSNs are in the superficial layer. The crypt OSNs reside in the superficial layer of the OE. The axons of these three types of OSNs project to different regions of the olfactory bulb, suggesting that these OSNs have distinct functions [28, 54]. We confirmed that OMP1 was mainly expressed in the deep layer of the OE (Fig. 6, Additional file 1: Figure S1), and was coexpressed with Gαolf2 (Additional file 3: Figure S3) but not with TRPC2 (Additional file 1: Figure S1). These results indicate that OMP1 corresponds to the previously characterized zebrafish OMP [27]. In contrast, OMP2 was expressed in the superficial layer (Figs. 5 and 6), in which the cell bodies of microvillous and crypt cells are situated. So, we initially considered that OMP2-expressing cells might be microvillous or crypt cells. However, this assumption seems unlikely because OMP2-expressing cells also express Gαolf2 (Fig. 7a-c), and neither microvillous nor crypt OSNs express Gαolf [54, 55]. Furthermore, we examined the coexpression of OMP2 with Ora genes [48, 49]. In particular, Ora4 is expressed in zebrafish crypt OSNs [56]. Although these genes were expressed in the OE, none of them was coexpressed with OMP2 (Additional file 4: Figure S4). Recently, a fourth type of OSN, kappe neuron, was identified, and these neurons are distributed in the superficial layer of the zebrafish OE [57]. They do, however, express Gαo [57]. Taken together, these results suggest that the OMP2/Gαolf2-coexpressing cells are most likely to be ciliated OSNs, in spite of the fact that cell bodies were distributed in the superficial layer. Probably, they also coexpress some ORs and/or TAARs. To definitively determine the cell type, a specific antibody against OMP2 is required. Nonetheless, the almost completely non-overlapping expression of OMP1 and 2 (Fig. 6a-c) implies that OMP1- and 2-expressing cells possess distinct roles in the OE.
Subfunctionalization between OMP1 and OMP2 in the olfactory system
OMP2 was expressed in the superficial layer of the OE (Figs. 6 and 7), whereas OMP1 was mainly expressed in the deep layer (Fig. 6, Additional file 1: Figure S1). The non-overlapping expression of OMP1 and 2 can be explained by subfunctionalization, which is a model for paralog retention attributed to the reduction and specialization of expression. The 3R derived paralogs of OMP in teleosts have partitioned their expression and perhaps function since the WGD event (Fig. 8, Additional file 5: Figure S5). Given that a single OMP gene is expressed in all of area of OE in mice and frogs, it is speculated that the single OMP gene was expressed in all Gαolf+ OSNs in the ancestral group (Additional file 5: Figure S5). In addition, because the OMP is known to play an important and fundamental role in signal transduction in OSNs [e.g. 29], OMP could be expressed in all OSNs of OE in the ancestral group that can be assessed by investigating the OE of non-teleost fish. We are now speculating that OMP could be expressed in all Gαolf+ OSNs in the spotted gar (Additional file 5: Figure S5, shown by blue characters). At present, however, the expression data of spotted gar was lacking, making it difficult to examine in this study.
By focusing on two OMPs in teleosts, we proposed the scenarios of subfunctionalization of 3R-derived paralogs. To further verify this scenarios, it is important to incorporate the information about the ancestral states, which are represented by extinct species or close relatives of teleosts. The basal lineages of ray-finned fish (non-teleost fish), which did not undergo the 3R, could be ideal species to infer the ancestral state. Accordingly, the expression pattern of OMP in gar and/or polypterus, should be analyzed in detail based on in situ hybridization etc. in the near future.
Other duplications of OMP in vertebrate evolution
African clawed frog has two OMPs in its genome [36]. Our phylogenetic analysis suggested that xenopus OMPs emerged in the African clawed frog lineage. The African clawed frog is an allotetraploid animal [58, 59], and the most recent WGD was estimated to have occurred ~30 MYA [58]. Accordingly, OMP duplication in African clawed frog is likely to be derived from a xenopus-specific WGD. Although the two xenopus OMPs show distinct expression patterns [36], such expression patterns do not appear to be mutually exclusive. Incomplete differentiation of xenopus OMPs is attributed to more recent WGD than 3R. Two OMPs are also present in the salmon genome [38]. Our results showed that both salmon OMPs are included in the OMP2 clade. It is well known that another round of WGD occurred independently in the salmon lineage [60, 61]. Therefore, it is most likely that two salmon OMP2s found in the present study emerged by this additional WGD. Interestingly, it has been suggested that certain groups of genes tend to be specifically retained after a WGD event [13, 61], and OMP would seem to be one of these genes. As salmon genome data become available, it will be interesting to attempt to locate OMP1 for further analysis.
Conclusions
We suggested that OMP paralogs, which were derived from 3R, have been retained in visual and olfactory system by subfunctionalization (Fig. 8). The expression pattern of OMP in gar or polypterus (ray-finned fish without 3R) should be investigated to confirm this scenarios in the future study. In addition, we propose that OMP2 could be used as a novel molecular marker of OSNs because OMP1 and 2 were separately expressed in the OE. Thus, the 3R-derived duplicated genes might become promising markers for the classification of various types of cells in the same organ, such as neural tissues.
Methods
Ethic statement
The animal protocols and procedures used in this study were approved by the Institutional Animal Care and Use Committee of Tokyo Institute of Technology [62].
Data mining
Human (Homo sapiens), mouse (Mus musculus), Western clawed frog, African clawed frog, zebrafish, and salmon OMP nucleotides sequences were acquired from DNA Data Bank of Japan (DDBJ) with the ARSA keyword search [63]. Accession numbers are as follows: human, BC069365; mouse, U02557; Western clawed frog, BC061304; African clawed frog, AJ010978, AJ010979; zebrafish, AF457189; salmon, AB490250, AB490251. These sequences were used as queries for a BLASTN search to acquire zebrafish, stickleback, and tilapia OMP cDNA sequences from the DDBJ EST database [64]. Accession numbers from the EST database are listed in Table 1. Other OMP sequences were acquired from Ensembl genome browser [65] with a TBLASTN search. For all analyses, a BLAST cutoff E-value was set at 1. Then, complete coding sequences were estimated by GeneWise [66]. The same method was used to acquire CAPN5 sequences. Accession numbers are as follows: human, BC018123; mouse, BC014767; Western clawed frog, BC075496; African clawed frog, BC048218. Information about OMP loci for syntenic analysis was also acquired from Ensembl with a BLASTN search.
Phylogenetic analysis
Deduced amino acid sequences of OMPs were aligned by ClustalW2 [67] with default parameters. Because of its low similarity, exon1 of OMP2 and the homologous regions of the other OMPs were removed from the alignment, and then the maximum likelihood phylogeny was constructed with MEGA6 [68] based on the multiple sequence alignment, using the amino acids WAG + F model with 10,000 bootstrap repetitions and other default parameters.
RT-PCR
The zebrafish were euthanized under anesthesia using ethyl 4-aminobenzoate. Total RNA was extracted from each organ of two adult zebrafish (strain Tü, 12–24 months old) with TRIzol (Invitrogen). After RNase-free DNase I (TaKaRa) digestion, each RNA sample was diluted to 10 ng/μl. cDNA was synthesized from 100 ng total RNA with SuperScript III Reverse Transcriptase (Invitrogen) using oligo-dT18 as a primer for 1 h at 50 °C. Genomic DNA for control was extracted from fins of adult zebrafish with DNeasy Blood & Tissue Kit (QIAGEN). PCR amplification was carried out for 30 s at 94 °C, 30 s at 55 °C, and 40 s at 72 °C for 35 cycles. Sequences of primers are listed in Table 2. To eliminate contamination of the PCR products derived from the genomic DNA, we designed intron-spanning primers for OMP2 and GAPDH.
Transcriptome data analysis
Transcriptome data from the eyes are acquired from DDBJ sequence read archive [69]. Accession numbers are as follows: spotted gar, SRR1288001 and SRR1288144; zebrafish, SRR1562528. Fragments per kilobase of exon per million mapped fragments (FPKM), which reflect relative expression level, were calculated by bowtie-2.2.5 [70] and rsem-1.2.21 [71].
Riboprobe synthesis
Each zebrafish RT-PCR product was ligated into pBluescript II SK(−) vector. Sequences of primers used for RT-PCR are listed in Table 2. Degenerate primers were designed to amplify several paralogs. After cloning and sequencing, the plasmids were extracted with the QIAfilter Plasmid Midi Kit (QIAGEN) and then linearized with the appropriate restriction enzyme. Digoxigenin (DIG)-labeled or fluorescein-labeled riboprobes were synthesized with T7 or T3 RNA polymerase (Roche) from the linearized plasmids with DIG or fluorescein RNA labeling mix (Roche), respectively. The riboprobes were treated with recombinant DNase I (TaKaRa) to exclude template plasmids.
Tissue preparation
Olfactory rosettes and eyes of adult zebrafish were dissected out, and fixed in 4 % paraformaldehyde (PFA) in phosphate-buffered saline (PBS) overnight at 4 °C. After fixation, tissues were cryoprotected in 20 % sucrose in PBS, embedded in O.C.T. compound (Sakura Finetek), and sectioned at a thickness of 10 μm on a cryostat (Leica). Sections were stored at −80 °C until use.
Fluorescence in situ hybridization
Sections were pretreated with 4 % PFA in PBS for 5 min, followed by treatment with 0.3 % H2O2 in PBS for 15 min and then with 5 μg/ml proteinase K in PBS for 10 min at 37 °C. After fixation with 4 % PFA in PBS for 10 min, sections were treated with 0.2 % glycine in PBS for 5 min, and with 0.2 N HCl for 20 min, followed by 0.25 % acetic anhydride/0.03 N HCl/0.1 M triethanolamine for 3 min. Sections were prehybridized with hybridization solution, which consisted of 50 % formamide; 10 mM Tris–HCl buffer, pH 7.5; 0.6 M NaCl; 1 mM EDTA; 0.25 % SDS; 1× Denhardt’s solution; 5 % dextran sulfate; and 0.2 mg/ml Yeast tRNA, for 40 minutes and were then hybridized with the hybridization solution containing 5 ng/μl DIG-labeled riboprobe at 60 °C overnight. After hybridization, sections were washed sequentially at 50 °C in 5× saline-sodium citrate (SSC), 50 % formamide in 5× SSC (twice), and then in 10 mM Tris–HCl, pH 7.5, containing 150 mM NaCl and 1 mM EDTA (TNE). After RNase treatment with 2 μg/ml RNase A in TNE for 30 min at 37 °C, sections were washed at 50 °C in 2× SSC (twice) and 0.2× SSC (twice). After treatment of the sections with streptavidin/biotin blocking kit (Vector Laboratories) and 1 % blocking reagent (PerkinElmer) in TBS, bound riboprobe was detected with peroxidase-conjugated anti-DIG antibody (1:100; Roche), and visualized with the TSA Plus biotin kit (PerkinElmer) and Alexa 594-conjugated streptavidin (1:500; Molecular Probes). Sections were coverslipped with VECTASHIELD mounting medium with 4’,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories), and images were digitally captured on a fluorescence microscope (Carl Zeiss). In the case of two-color detection, fluorescein-labeled riboprobe was mixed with DIG-labeled riboprobe, and used for hybridization. Fluorescein-labeled riboprobe was detected with peroxidase-conjugated anti-fluorescein antibody (1:500; PerkinElmer), and was visualized with the TSA Plus 2,4-dinitrophenyl (DNP) system (PerkinElmer) and Alexa 488-conjugated anti-DNP antibody (1:500; Molecular Probes). After the detection of the fluorescein-labeled riboprobe, sections were treated with 15 % H2O2 in PBS for 30 min to inactivate peroxidase. Then, the DIG-labeled riboprobe was detected as described above.
Availability of data and materials
The data sets supporting the results of this article are available as Additional file.
Abbreviations
- WGD:
-
Whole genome duplication
- OE:
-
Olfactory epithelium
- OMP:
-
Olfactory marker protein
- OSN:
-
Olfactory sensory neuron
- KO:
-
Knockout
- OR:
-
Odorant receptor
- TAAR:
-
Trace amine-associated receptor
- V1R:
-
Vomeronasal type 1 receptor
- V2R:
-
Vomeronasal type 2 receptor
References
Ohno S. Evolution by gene duplication. New York: Springer; 1970.
Zhang J, Rosenberg HF, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci U S A. 1998;95:3708–13.
Zhang J. Evolution by gene duplication: An update. Trends Ecol Evol. 2003;18:292–8.
Holland PW, Garcia-Fernàndez J, Williams NA, Sidow A: Gene duplications and the origins of vertebrate development. Dev Suppl 1994, 125–133.
Kasahara M. The 2R hypothesis: An update. Curr Opin Immunol. 2007;19:547–52.
Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–71.
Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45:415–21.
Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y. Major events in the genome evolution of vertebrates: Paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci U S A. 2004;101:1638–43.
Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–4.
Meyer A, Schartl M. Gene and genome duplications in vertebrates: The one-to-four (−to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;11:699–704.
Hoegg S, Meyer A. Hox clusters as models for vertebrate genome evolution. Trends Genet. 2005;21:421–4.
Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol. 2004;59:190–203.
Sato Y, Hashiguchi Y, Nishida M. Temporal pattern of loss/persistence of duplicate genes involved in signal transduction and metabolic pathways after teleost-specific genome duplication. BMC Evol Biol. 2009;9:127–7.
Takamatsu N, Kurosawa G, Takahashi M, Inokuma R, Tanaka M, Kanamori A, et al. Duplicated Abd-B class genes in medaka hoxAa and hoxAb clusters exhibit differential expression patterns in pectoral fin buds. Dev Genes Evol. 2007;217:263–73.
Hurley IA, Scemama JL, Prince VE. Consequences of hoxb1 duplication in teleost fish. Evol Dev. 2007;9:540–54.
Davis A, Stellwag EJ. Spatio-temporal patterns of Hox paralog group 3–6 gene expression during Japanese medaka (Oryzias latipes) embryonic development. Gene Expr Patterns. 2010;10:244–50.
Lyon RS, Davis A, Scemama JL. Spatio-temporal expression patterns of anterior Hox genes during Nile tilapia (Oreochromis niloticus) embryonic development. Gene Expr Patterns. 2013;13:104–8.
Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–45.
Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–73.
Rastogi S, Liberles DA. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol. 2005;5:28–8.
Kleinjan DA, Bancewicz RM, Gautier P, Dahm R, Schonthaler HB, Damante G, et al. Subfunctionalization of duplicated zebrafish pax6 genes by cis-regulatory divergence. PLoS Genet. 2008;4:e29.
Margolis FL. A brain protein unique to the olfactory bulb. Proc Natl Acad Sci U S A. 1972;69:1221–4.
Farbman AI, Margolis FL. Olfactory marker protein during ontogeny: Immunohistochemical localization. Dev Biol. 1980;74:205–15.
Danciger E, Mettling C, Vidal M, Morris R, Margolis F. Olfactory marker protein gene: Its structure and olfactory neuron-specific expression in transgenic mice. Proc Natl Acad Sci U S A. 1989;86:8565–9.
Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, et al. Visualizing an olfactory sensory map. Cell. 1996;87:675–86.
Potter SM, Zheng C, Koos DS, Feinstein P, Fraser SE, Mombaerts P. Structure and emergence of specific olfactory glomeruli in the mouse. J Neurosci. 2001;21:9713–23.
Celik A, Fuss SH, Korsching SI. Selective targeting of zebrafish olfactory receptor neurons by the endogenous OMP promoter. Eur J Neurosci. 2002;15:798–806.
Sato Y, Miyasaka N, Yoshihara Y. Mutually exclusive glomerular innervation by two distinct types of olfactory sensory neurons revealed in transgenic zebrafish. J Neurosci. 2005;25:4889–97.
Buiakova OI, Baker H, Scott JW, Farbman A, Kream R, Grillo M, et al. Olfactory marker protein (OMP) gene deletion causes altered physiological activity of olfactory sensory neurons. Proc Natl Acad Sci U S A. 1996;93:9858–63.
Youngentob SL, Margolis FL. OMP gene deletion causes an elevation in behavioral threshold sensitivity. NeuroReport. 1999;10:15–9.
St John JA, Key B. Olfactory marker protein modulates primary olfactory axon overshooting in the olfactory bulb. J Comp Neurol. 2005;488:61–9.
Lee AC, He J, Ma M. Olfactory marker protein is critical for functional maturation of olfactory sensory neurons and development of mother preference. J Neurosci. 2011;31:2974–82.
Kwon HJ, Koo JH, Zufall F, Leinders-Zufall T, Margolis FL. Ca extrusion by NCX is compromised in olfactory sensory neurons of OMP mice. PLoS One. 2009;4:e4260.
Buiakova OI, Krishna NS, Getchell TV, Margolis FL. Human and rodent OMP genes: Conservation of structural and regulatory motifs and cellular localization. Genomics. 1994;20:452–62.
Smith PC, Firestein S, Hunt JF. The crystal structure of the olfactory marker protein at 2.3 A resolution. J Mol Biol. 2002;319:807–21.
Rössler P, Mezler M, Breer H. Two olfactory marker proteins in Xenopus laevis. J Comp Neurol. 1998;395:273–80.
Yasuoka A, Endo K, Asano-Miyoshi M, Abe K, Emori Y. Two subfamilies of olfactory receptor genes in medaka fish, Oryzias latipes: Genomic organization and differential expression in olfactory epithelium. J Biochem. 1999;126:866–73.
Kudo H, Doi Y, Ueda H, Kaeriyama M. Molecular characterization and histochemical demonstration of salmon olfactory marker protein in the olfactory epithelium of lacustrine sockeye salmon (Oncorhynchus nerka). Comp Biochem Physiol A Mol Integr Physiol. 2009;154:142–50.
ZFIN: The Zebrafish Model Organism Database [http://zfin.org]
Long EO, David ID. Repeated genes in eukaryotes. Annu Rev Plant Physiol Plant Mol Biol. 1980;49:727–64.
Yazulla S, Studholme KM. Neurochemical anatomy of the zebrafish retina as determined by immunocytochemistry. J Neurocytol. 2002;30:551–92.
Buck L, Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65:175–87.
Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–50.
Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206.
Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–73.
Matsunami H, Buck LB. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90:775–84.
Oka Y, Korsching SI. Shared and unique G alpha proteins in the zebrafish versus mammalian senses of taste and smell. Chem Senses. 2011;36:357–65.
Saraiva LR, Korsching SI. A novel olfactory receptor gene family in teleost fish. Genome Res. 2007;17:1448–57.
Ota T, Nikaido M, Suzuki H, Hagino-Yamagishi K, Okada N. Characterization of V1R receptor (ora) genes in Lake Victoria cichlids. Gene. 2012;499:273–9.
Pronin A, Levay K, Velmeshev D, Faghihi M, Shestopalov VI, Slepak VZ. Expression of olfactory signaling genes in the eye. PLoS One. 2014;9:e96435.
Langenbacher AD, Dong Y, Shu X, Choi J, Nicoll DA, Goldhaber JI, et al. Mutation in sodium-calcium exchanger 1 (NCX1) causes cardiac fibrillation in zebrafish. Proc Natl Acad Sci U S A. 2005;102:17699–704.
Liao BK, Deng AN, Chen SC, Chou MY, Hwang PP. Expression and water calcium dependence of calcium transporter isoforms in zebrafish gill mitochondrion-rich cells. BMC Genomics. 2007;8:354–4.
Hansen A, Zeiske E. The peripheral olfactory organ of the zebrafish, Danio rerio: An ultrastructural study. Chem Senses. 1998;23:39–48.
Hansen A, Rolen SH, Anderson K, Morita Y, Caprio J, Finger TE. Correlation between olfactory receptor cell type and function in the channel catfish. J Neurosci. 2003;23:9328–39.
Hansen A, Anderson KT, Finger TE. Differential distribution of olfactory receptor neurons in goldfish: Structural and molecular correlates. J Comp Neurol. 2004;477:347–59.
Oka Y, Saraiva LR, Korsching SI. Crypt neurons express a single V1R-related ora gene. Chem Senses. 2012;37:219–27.
Ahuja G, Nia SB, Zapilko V, Shiriagin V, Kowatschew D, Oka Y, et al. Kappe neurons, a novel population of olfactory sensory neurons. Sci Rep. 2014;4:4037–7.
Bisbee CA, Baker MA, Wilson AC, Haji-Azimi I, Fischberg M. Albumin phylogeny for clawed frogs (Xenopus). Science. 1977;195:785–7.
Jeffreys AJ, Wilson V, Wood D, Simons JP, Kay RM, Williams JG. Linkage of adult alpha- and beta-globin genes in X. laevis and gene duplication by tetraploidization. Cell. 1980;21:555–64.
Moghadam HK, Ferguson MM, Danzmann RG. Evidence for Hox gene duplication in rainbow trout (Oncorhynchus mykiss): A tetraploid model species. J Mol Evol. 2005;61:804–18.
Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, Noël B, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014;5:3657–7.
Hau J, van Hoosier GL. Hand book of laboratory animal science, Volume I: Essential Principles and Practices. 2nd ed. London: CRC Press; 2002.
DNA Data Bank of Japan ARSA search [http://ddbj.nig.ac.jp/arsa/]
DNA Data Bank of Japan BLAST search [http://blast.ddbj.nig.ac.jp/]
Ensembl Genome Browser [http://www.ensembl.org/index.html]
GeneWise [http://www.ebi.ac.uk/Tools/psa/genewise/]
ClustalW2 [http://www.clustal.org/clustal2/]
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2014;30:2725–9.
DDBJ sequence read archive [http://trace.ddbj.nig.ac.jp/dra/]
Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323–3.
Acknowledgments
We thank Nobuko Yamada for maintaining the fish. We also thank Yoshihiro Yoshihara for meaningful discussions about this work. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan 21227002 (NO), and JSPS Asia-Africa Science Platform Program (NO).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HS participated in the design of the study, carried out analyses, and drafted the manuscript. MN initiated the study, participated in its design, and helped to draft the manuscript. KH technically supported the histological analyses. NO participated in the design of the study, supervised the study and finalized the manuscript. All authors read and approved the final manuscript.
Additional files
Additional file 1: Figure S1.
Expression patterns of OMP1 and TRPC2. (PDF 4.99 mb)
Additional file 2: Figure S2.
Expression patterns of Gαolf2 and Gαo2. (PDF 4.99 mb)
Additional file 3: Figure S3.
Expression patterns of OMP1 and Gαolf2. (PDF 4.99 mb)
Additional file 4: Figure S4.
Expression patterns of OMP2 and Ora genes. (PDF 4.99 mb)
Additional file 5: Figure S5.
Schematics of OMP duplication and subdivision of gene expressions during evolution. (PDF 4.99 mb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Suzuki, H., Nikaido, M., Hagino-Yamagishi, K. et al. Distinct functions of two olfactory marker protein genes derived from teleost-specific whole genome duplication. BMC Evol Biol 15, 245 (2015). https://doi.org/10.1186/s12862-015-0530-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-015-0530-y