Abstract
Background
Rice blast (causative pathogen Magnaporthe oryzae) represents a major biotic constraint over rice production. While numerous genes for resistance have been found in both japonica and indica germplasm, as yet the diversity harbored by aus germplasm has not been widely exploited.
Results
The blast resistance present in the aus type cultivar AS20-1 was shown, via an analysis of segregation in the F2 generation bred from a cross with the highly blast susceptible cultivar Aichi Asahi, to be due to the action of a single recessive gene, denoted pi66(t). The presence of pi66(t) gave an intermediate level control to plants infected with the blast pathogen isolate EHL0635. A bulked segregant analysis indicated that four microsatellite loci (SSRs) mapping to chromosome 3 were probably linked to pi66(t). Localized mapping using chromosome 3-based SSRs and Indels defined a genetic window for pi66(t), flanked by the markers F04-j2 and M19-i12, which physically equals to 27.7 and 49.0 kb, respectively, in the reference genomes of cultivars Nipponbare and 93–11. This physical interval does not harbor any major gene currently associated with disease resistance.
Conclusion
pi66(t) is one of just three recessive genes controlling rice blast, and is the first major gene for resistance to be mapped to chromosome 3.
Similar content being viewed by others
Background
Rice, a crop which feeds half of the world’s population, has been cultivated for at least 8,000 years (Khush 1997; The 3,000 rice genome project 2014; Travis et al. 2015). Five distinct groups of rice germplasm have long been recognized: they are referred to as indica, aus, basmati/sadri, tropical japonica and temperate japonica (The 3,000 Rice Genomes Project 2014; Travis et al. 2015). The aus group has developed in the north-eastern region of the Indian sub-continent, where both the climate and the growing environment are highly variable (Mahender et al. 2012; Travis et al. 2015). In recent years, aus germplasm has grown in importance as a source of genes for rice improvement, especially in the context of breeding for resistance/tolerance to abiotic and biotic stress (Travis et al. 2015 and references therein).
Rice blast (causative pathogen Magnaporthe oryzae) is a major constraint over rice production, inducing grain yield losses of up to 90 % (He et al. 2012; Singh et al. 2015). Although breeders have so far been able to rely on a number of sources of genetic resistance, the pathogen is adept at evolving new races, with the result that mongenic resistances typically break down quite rapidly (Wu et al. 2014; Singh et al. 2015; Zhang et al. 2015). To date, some one hundred rice blast resistance (Pi) genes have been identified, many of which have been shown to map within a cluster or even in form of a tandem array; they are dispersed on eleven of the twelve rice chromosomes (Sharma et al. 2012; Singh et al. 2015; Tanwaeer et al. 2015 and references therein). All but two of the Pi genes are functionally dominant (Fukuoka et al. 2009; He et al. 2012), and about 30 have been isolated: their products mostly belong to the large group of nucleotide-binding site (NBS)-leucine-rich repeat (LRR) proteins. The two exceptions are Pid-2 and pi21 (Chen et al. 2006b; Fukuoka et al. 2009; Liu et al. 2011). Here, a third recessive gene, denoted pi66(t), has been identified in the aus cultivar (cv.) AS20-1, and its genomic position has been defined.
Results
Resistance Reaction and Spectrum
Numerous differential reactions were identified among the four cvs in the five Mo populations, suggesting that the Pi gene(s) carried by the donor cv. AS20-1 could be distinguished from the other Pi genes with these reactions (Table 1 and Additional file 1: Table S1). Intermediate and even lower resistance frequencies were evaluated among the four cvs in the five M. oryzae populations, indicating that all the four Pi genes should be incorporated with other Pi genes to stand the higher level of resistance in a given cultivar, if it will be released in the five M. oryzae populations.
Resistance Inheritance
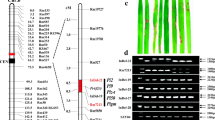
When challenged by the blast isolate EHL0635, cv. AS20-1 was scored as moderately resistance (MR), cv. Aichi Asahi as susceptible (S) and the cv. AS20-1 x cv. Aichi Asah F1 as highly susceptible (HS) (Fig. 1). The qPCR-based assay confirmed that the hybrid was more susceptible than cv. Aichi Asahi. The F2 progeny segregated as 101 R, 282 MR, 254 MS and 883 S, fitting a monogenic 1R:3S ratio when the R/MR and MS/S classes were combined (χ 2 = 0.02; P > 0.80; Table 2). Together, these results indicated that the blast resistance expressed by cv. AS20-1 relied on homozygosity for the recessive allele of a single gene.
The effectiveness of pi66(t) to resist infection by M. oryzae. a The infection phenotype of cvs AS20-1 (resistant), Aichi Asahi (susceptible) and the cv. AS20-1 x cv. Aichi Asahi F1 hybrid (highly susceptible). b qPCR-based quantification of infection. Each bar represents the mean ± standard deviation (n = 3). Similar results were obtained from two biological replications each with three technical repeats. W: mock inoculation with water, P: inoculation with the pathogen isolate EHL0635
Gene Locus
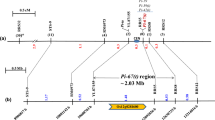
BSA analysis revealed that four SSR markers (RM487, RM16, RM55, and RM168) on rice chromosome 3 were candidate markers linking to the target Pi gene, exclusively, in the F2 population. The first round of linkage analysis with 750 viable F2 plants revealed that there were 64 and 37 recombinants, respectively, at RM487 and RM16 loci on the centromere side, 35 and 22 distinct recombinants, respectively, at RM168 and RM55 loci on the telomere side, indicating that the four candidate markers were indeed linkage markers with the target Pi gene (Fig. 2a). Because no major Pi gene had been previously identified in this region, the novel Pi gene in AS20-1 was designated as pi66(t).
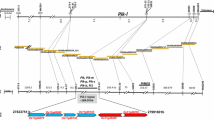
The genomic location of pi66(t). a Physical maps of the pi66 region based on the reference sequences of cvs 93–11 and Nipponbare. The numbers shown below the map represent the physical distance in kb, those shown in parentheses represent the numbers of recombinants/gametes detected in the mapping population. b A BAC contig map of the pi66(t) region derived from the cv. Nipponbare tiling map. c The predicted gene content of the mapping interval harboring pi66(t). Three substantial Indel events, which results in six genome-specific genes presented in the region, were determined via genome comparison and P/A analyses, of which three chimeric genes presented in both 93–11 and AS20-1 genomes. Candidate genes of cvs Nipponbare, 93–11, and As20-1 were indicated with grey, black, and blank arrows, respectively
Additional nine polymorphic SSR markers developed in the region defined by the flanking markers RM16 and RM55 were subjected to the second round of linkage analysis (Additional file 2: Table S2). The results showed that there were 31 to 22 recombinants detected among the seven marker loci [B07 (31), G02 (30), H15 (30), N03 (30), P23 (30), N11 (29), L18 (25), M23 (22)] on the centromere side, and only 8 distinct recombinants at RM135 locus on the telomere side (Fig. 2a). A total of 14 additional Indel markers developed in the narrower region flanked by markers M23 and RM135 were subjected to the third round of linkage analysis (Additional file 2: Table S2). The results showed that there were 15 to 2 recombinants detected among the six marker loci [D21 (15), E06 (9), I20 (7), I24 (2), F04 (2), F04-j2 (2)] on the centromere side, and 7 to 1 recombinant(s) detected among the eight marker loci [G23 (7), E01 (4), M19 (3), M19-4 (3), M19-3 (2), M19-2 (2), M19-1 (1), M19-i12 (1)] on the telomere side (Fig. 2a). The target locus, pi66(t), was closely flanked by F04-j2 and M19-i12, which equals to 27.7 and 49.0 kb, respectively, in the reference genomes of cvs Nipponbare and 93–11 (Fig. 2a).
Candidate Genes
The pi66(t) region was represented by the two cv. Nipponbare overlapping BACs OSJNBb0009F04 and OSJNBa0092M19 (Fig. 2b). The number of genes present within this region was six in cv. Nipponbare and 14 in cv. 93–11 (Additional file 3: Table S3). Genome comparison and presence/absence (P/A) analyses revealed that there were three substantial Indel events that resulted in six genome-specific genes in the region. That is, both pi66-2j and pi66-3j in Indel I present in two genomes of cvs Nipponbare and AS20-1; pi66-1i-2 (a duplication of pi66-1i-1) in Indel II, pi66-5i and pi66-6i in Indel III in that of both cvs 93–11 and AS20-1; and pi66-2i in Indel II in that of cv 93–11, only (Fig. 2c, Additional file 4: Figure S1). Notably, there were six transposon-like genes (pi66-1j, -2j, -3j, 1i-1, 1i-2, -6i), of which both pi66-2j and -3j were scattered across the entire genomes except for the target region of cv. 93–11, thereby ruling out for P/A analyis (Additional file 4: Figure S1; Additional file 3: Table S3). Furthermore, there were three chimeric genes in both 93–11 and AS20-1 genomes (Fig. 2c and Additional file 4: Figure S1). By excluding six transposon-like genes, there were three most possible candidates (pi66-5a, -6a, -7a) for pi66(t) (Fig. 2c; Additional file 3: Table S3).
Discussion
Chinese rice breeders have to date largely ignored aus germplasm, even though it has acquired a growing reputation for harboring genes for resistance/tolerance to abiotic and biotic stress (Travis et al. 2015). Rather, efforts to improve indica have concentrated on materials developed in SE Asia, while those directed at japonica have relied on germplasm from Japan (Wu et al. 1991). In addition to pi66(t), aus germplasm has also yielded both Pi16 and an allele of Pik (Pan et al. 1999). More recently, nine already recognized Pi genes have been identified as present in materials originating in NE and E India (Imam et al. 2014), which is the center of origin of aus germplasm. Notably, the donor of pi66(t) also harbors a gene conferring resistance against the brown plant hopper; this gene also lies on chromosome 3, but at some distance from pi66(t) (Chen et al. 2006a). Such works clearly indicated that aus cvs are valuable and promising genetic resources for withstanding biotic pressures including rice blast disease, and will greatly enlarge the gene pool for rice breeders.
Plant disease resistance genes have been classified into two types, the most frequent of which encode an NBS-LRR protein. Non-NBS-LRR genes encode a wide diversity of products (Chauhan et al. 2015; Olukolu et al. 2016), tend to confer partial (rather than complete) resistance and are typically more durable than the NBS-LRR type genes. The most well documented non-NBS-LRR type is barley mlo, a gene which encodes a G protein-coupled receptor residing in the plasma membrane (Kim et al. 2002); the gene confers durable resistance to a broad spectrum of powdery mildew races (Acevedo-Garcia et al. 2014). A second example is the wheat gene Lr34, which encodes an ATP-binding cassette transporter; its product protects against five distinct foliar fungal pathogens (Kratttinger et al. 2009; Chauhan et al. 2015). The maize gene ZmWAK encodes a plasma membrane-related receptor-like kinase; its presence has been correlated with a reduction in the incidence of head smut disease (Zuo et al. 2015). Finally, the rice gene xa5 encodes a small subunit of the transcription factor IIA (TFIIA); this gene confers resistance against bacterial blight (Iyer-Pascuzzi 2004). Before the identification of pi66(t), all but two of the Pi genes characterized to date act as dominant alleles. The exceptions are pi21 and pi55. The former gene encodes a proline-rich protein harboring a probable heavy metal-binding domain and some predicted protein-protein interaction motifs; the resistant allele differs from the wild type dominant one by two deletions affecting the latter motifs, and which are thought to be responsible for the allele's determination of non-race-specific resistance (Fukuoka et al. 2009). One of candidate genes for pi55 encodes a protein rather similar to that encoded by pi21. Although a substantial number of major Pi genes have been intra-chromosomally mapped, pi66(t) is the first to be located on chromosome 3. Two quantitative trait loci mapping to this chromosome (Os03g0122000 and Os03g0120400) have been associated with blast resistance (Wang et al. 2014), but both lie outside the critical RM16-RM55 interval. The pi66 identified in the current study that is the third recessive Pi gene located on the virgin land, where no any known Pi protein (domain) is identifiable (Additional file 3: Table S3). It is noteworthy that the bph19 derived from the donor cv. AS20-1 was also recognized as non-NBS-LRR resistance gene (Chen et al. 2006a). It has been argued that durable and broad-spectrum resistance may be more readily achieved by deploying non-NBS-LRR genes, perhaps in combination with NBS-LRR ones, than by attempting to stack genes which each (at least for some time) confer immunity (Fukuoka et al. 2009; Acevedo-Garcia et al. 2014; Chauhan et al. 2015; Zuo et al. 2015). This hypothesis can only be tested by exploiting genes such as pi66(t) in a rice breeding program.
Conclusions
This research has confirmed that novel resistance genes against blast can be recovered from aus germplasm. The gene pi66(t) identified here is the third recessive Pi gene to be identified, and is also the first major Pi gene to be located on chromosome 3.
Methods
Phenotyping
The pi66(t) donor cv. AS20-1, along with the Pia carrier cv. Aichi Asahi, the Pi36 carrier cv. Kasalath and the Pita-2 carrier cv. IRBLta2-Pi were challenged with 60 M. oryzae isolates collected from each of Guangdong (GD), Guangxi (GX), Yunnan (YN), Sichuan (SC) and Heilongjiang (HLJ) provinces. Inoculation and scoring methods were adapted from those described by Pan et al. (1996, 2003). Plants were assigned a score of either 0–1 (resistant: R), 2–3 (moderately resistant: MR), 4 (moderately susceptible: MS) or 5 (susceptible: S). The frequency of resistance for each of the four cultivars within each of the five M. oryzae populations was calculated from [(R + MR)/(R + MR) + (MS + S)]. The typical reactions of cv. AS20-1, cv. Aichi Asahi and the cv. AS20-1 x cv. Aichi Asahi F1 plants were quantified using a quantitative PCR (qPCR) assay, according to the protocols previously described (Berruyer et al. 2006; Kawano et al. 2010; Zhang et al. 2015). Oryza sativa OsUbi (Gene ID: 4332169) and M. oryzae Pot2 (Gene ID: 2680652) were used as the reference genes for, respectively, the host and the pathogen DNA.
Chromosome Mapping
The donor cv. AS20-1 was crossed with the highly susceptible cv. Aichi Asahi, and their F2 progenies were screened for reaction to inoculation with M. oryzae isolate EHL0635. The F2 population showing monogenic segregation was regarded as the mapping population, thereby subjecting to the bulked-segregant assay (BSA) for quickly mapping chromosomal region involving the target gene. Genomic DNAs of the F2 plants as well as the parental plants were extracted from frozen leaves using the CTAB method. Two contrast bulks that were constructed by pooling equimolar amounts of DNAs from 10 resistant or 10 susceptible F2 plants. The two bulks, along with both parental DNAs, were then assayed with a set of 180 simple sequence repeat (SSR) markers (Temnykh et al. 2000, 2001), selected to span the full rice genome, following the methods given by He et al. (2012).
Gene Mapping
Genomic map of target gene were established through three rounds of linkage analysis using genomic position-ready molecular markers (He et al. 2012). The first round was carried out with candidate markers defined by BSA for screening recombinants on both sides of the target locus. The second round was carried out with additional SSR markers in the target region flanked by the closest markers derived from the first round of linkage analysis, which were developed on the basis of reference sequence of cv. Nipponbare, except for RM135 that was adopted from the rice SSR marker maps (Temnykh et al. 2000, 2001). The third round was carried out in the recombinant progeny with insertion/deletion (Indel) markers those were developed de novo based on differential sequences between the two reference sequences of japonica cv. Nipponbare and indica cv. 93–11. Linkage marker search and prime designation were performed in the way essentially same way as previously described (Liu et al. 2005; Zeng et al. 2011; He et al. 2012). Genomic map of the target locus was constructed on the basis of both reference sequences.
Candidate Gene Indentification
Candidates for pi66(t) were predicted based on gene annotations provided by BLASTN (www.ncbi.nlm.nih.gov/BLAST), RiceGAAS (ricegaas.dna.affrc.go.jp) and FGENSH (www.softberry.com) software. The two reference sequences proved to be rather diverse in the target region, so candidates that encode proteins with over 200 aa was validated by PCR-based presence/absence (P/A) test against the four DNAs of cvs AS20-1, Aichi Asahi, 93–11, and Nipponbare, following Zhai et al. (2011).
References
Acevedo-Garcia J, Kusch S, Panstruga R (2014) Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol 204:273–281
Berruyer R, Poussier S, Kankanala P, Mosquera G, Valant B (2006) Quantitative and qualitative influence of inoculation methods on in planta growth of rice blast fungus. Phytopathology 96:346–355
Chauhan H, Boni R, Bucher R, Kuhn B, Buchmann G, Sucher J, Selter L, Hensel G, Kumlehn J, Bigler L, Glauser G, Wicker T, Krattinger S, Keller B (2015) The wheat resistance gene Lr34 results in the constitutive induction of multiple defense pathoways in transgenic barley. Plant J 84:202–215
Chen J, Wang L, Pan X, Pan Q (2006a) Genetic analysis and fine mapping of a rice brown planthopper (Nilaparvara lugens Stål) resistance gene bph19(t). Mol Gen Genomics 275:321–329
Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Ma B, Wang Y, Zhao X, Li S, Zhu L (2006b) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46:794–804
Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K, Yano M (2009) Loss of function of a proline-contaning protein confers durable disease resistance in rice. Science 325:998–1001
He X, Liu X, Wang L, Wang L, Lin F, Cheng Y, Chen Z, Liao Y, Pan Q (2012) Identification of the novel recessive gene pi55(t) conferring resistance to Magnaporthe oryzae. Sci China Life Sci 55(2):141–9
Imam J, Alam S, Mandal N, Variar M, Shukla P (2014) Molecular screening for identification of blast resistance genes in North East and Eastern Indian rice germplasm (Oryza sativa L.) with PCR based markers. Euphytica 196:199–211
Iyer-Pascuzzi MCS (2004) The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol Plant Microbe Interact 17:1348–1354
Kawano Y, Akamatsu A, Hayashi K, Housen Y, Okuda J, Yao A, Nakashima A, Takahashi H, Yoshida H, Wong H, Kawasaki T, Shimamoto K (2010) Activation of Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Mirobe 7:362–375
Khush G (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35:25–34
Kim M, Panstruga R, Elliott C, Müller J, Devoto A, Yoon H, Park H, Cho M, Schulze-Lefert P (2002) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416:447–451
Kratttinger S, Lagudah E, Spielmeyer W, Singh R, Huerta-Espino J, McFadden H, Bossolini E, Selter L, Keller B (2009) A putative ABC transporter confers durable resistance to multiple fungal pathongens in wheat. Science 323:1360–1363
Liu X, Wang L, Chen S, Lin F, Pan Q (2005) Genetic and physical mapping of Pi36(t), a novel rice blast resistance gene located on rice chromosome 8. Mol Gen Genomics 274:394–401
Liu X, Wei J, Zhang J, Wang C, Liu X, Zhang X, Wang L, Pan Q (2011) Genetic variation of rice blast resistance genes in Oryza sativa and its wild relatives. Int J Plant Sci 172:970–979
Mahender A, Swain D, Gitishree D, Subudhi H, Rao G (2012) Molecular analysis of native Manipur rice accessions for resistance against blast. African J Biotech 11:1321–1329
Pan Q, Hu Z, Tanisaka T, Wang L (2003) Fine mapping of the blast resistance gene Pi15, linked to Pii, on rice chromosome 9. J Integr Plant Biol (formerly Acta Bot Sin) 45:871–877
Pan Q, Wang L, Tanisaka T (1999) A new blast resistance gene identified in the Indian native rice cultivar Aus373 through allelism and linkage tests. Plant Pathol 48:289–293
Pan Q, Wang L, Ikehashi H, Tanisaka T (1996) Identification of a new blast resistance gene in the indica rice cultivar Kasalath using Japanese differential cultivars and isozyme markers. Phytopathology 86:1071–1075
Olukolu B, Tracy W, Wisser R, De Vries B, Balint-Kurti P (2016) A genome-wide association study for partial resistance to maize common rust. Phytopathology 106:745–751
Sharma T, Rai A, Gupta S, Vijayan J, Devanna B, Ray S (2012) Rice blast management through host-plant resistance: Retrospect and prospects. Agr Res 1:37–52
Singh W, Kapila R, Sharma T, Rathour R (2015) Genetic and physical mapping of a new allele of Pik locus from japonica rice ‘Lijiangxintuanheigu’. Euphytica 205:889–901
Tanwaeer F, Rafii M, Sijam K, Rahim H, Ahmed F, Latif M (2015) Current advance methods for the identification of blast resistance genes in rice. Comptes Rendus Biol 338:321–334
Temnykh S, Park W, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho Y, Ishii T, McCouch S (2000) Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor Appl Genet 100:697–712
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
The 3,000 Rice Genomes Project (2014) The 3,000 rice genomes project. GigaSci 3:7
Travis A, Norton GJ, Datta S, Sarma R, Dasgupta T, Savio F, Macaulay M, Hedley P, McNally K, Sumon M, Islam R, Price A (2015) Assessing the genetic diversity of rice originating from Bangladesh, Assam and West Bengal. Rice 8:35
Wang C, Yang Y, Yuan X, Xu Q, Feng Y, Yu H, Wang Y (2014) Genome-wide association study of blast resistance in indica rice. BMC Plant Biol 14:311
Wu M, Yin C, Liang N (1991) Rice germplasm. In: Lin S, Min S (eds) Rice varieties and their genealogy in China. Shanghai Scientific and Technical Publishers, Shanghai (In Chinese)
Wu W, Wang L, Zhang S, Li Z, Zhang Y, Lin F, Pan Q (2014) Stepwise arms race between AvrPik and Pik alleles in the rice blast pathosystem. Mol Plant Microbe Interact 27:759–769
Zeng X, Yang X, Zhao Z, Lin F, Wang L, Pan Q (2011) Characterization and fine mapping of the rice blast resistance gene Pia. Sci China Life Sci 54:372–378
Zhai C, Lin F, Dong Z, He X, Yuan B, Zeng X, Wang L, Pan Q (2011) The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol 189:321–334
Zhang S, Wang L, Wu W, He L, Yang X, Pan Q (2015) Function and evolution of Maganporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci Rep 5:11642
Zuo W, Chao Q, Zhang N, Ye J, Tan G, Li B, Xing Y, Zhang B, Liu H, Fengler K, Zhao J, Chen Y, Lai J, Yan J, Xu M (2015) A Maize wall-associated kinase confers quantitative resistance to head smut. Nat Genet 47:151–157
Acknowledgments
We acknowledge funding from the National Key R&D Project (2016YDF0100601), the National Transgenic Research Project (2015ZX08001-002), and the National Natural Science Foundation of China (U1131003). Materials used in the current study were obtained from our own collection with great helps from Dr. X. Zhu at the Guangdong Academy of Agricultural Sciences, Dr. R. Li at Guangxi University, Dr. Y. Zhu at Yunnan Agricultural University, Dr. D. Lu at Sichuan Academy of Agricultural Sciences, and Ms. Y. Zhang at Heilongjiang Bayi Agricultural University.
Competing Interests
The authors declare that they have no competing interests.
Authors’ Contributions
Project conception (QP). Resistance phenotyping (QP, LW). Mapping population construction (QP, LW). Linkage analysis (ZL, QP), Candidate gene analysis (ZL, QP). Manuscript preparation (QP). All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional Files
Additional file 1: Table S1.
Sampling and phenotyping information for the 300 isolates selected from five Chinese M. oryzae populations. (XLSX 31 kb)
Additional file 2: Table S2.
PCR-based markers mapping to the pi66(t) region. (DOCX 50 kb)
Additional file 3: Table S3.
The gene content of the region flanked by the pi66-linked markers F04-j2 and M19-i12. (DOCX 22 kb)
Additional file 4: Figure S1.
Presence/absence analysis of nine candidate genes. The upper panel shows the schematic gene structure used for primer design, and the lower two panels show the amplicons. 93: cv. 93–11, Ni: cv. Nipponbare, AS: cv. AS20-1, Ai: cv. Aichi Asahi, M1: size marker DL15,000; M2: size marker DL2,000. (PPTX 680 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Liang, Z., Wang, L. & Pan, Q. A New Recessive Gene Conferring Resistance Against Rice Blast. Rice 9, 47 (2016). https://doi.org/10.1186/s12284-016-0120-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-016-0120-7