Abstract
Background
Members of the genus Acanthamoeba are widely distributed throughout the world, and some of them are considered pathogenic, as they are capable of causing corneal and central nervous system diseases. In this study, we isolated Acanthamoeba strains from soil and tap water in Yanji, China.
Methods
We identified four strains of Acanthamoeba (CJY/S1, CJY/S2, CJY/S3, and CJY/W1) using mitochondrial DNA restriction fragment length polymorphism (mtDNA RFLP) analysis. Nuclear 18S rDNA sequences were used for phylogenetic analysis and species identification.
Results
Genotypic characterization of the isolates showed that they belonged to genotypes T4 (CJY/S1 and CJY/S2), T5 (CJY/S3), and T16 (CJY/W1). Sequence differences between CJY/S1 and Acanthamoeba castellanii Neff, CJY/S2 and Acanthamoeba KA/E7, and CJY/S3 and Acanthamoeba lenticulata 68–2 were 0.31, 0.2, and 0.26%, respectively. 18S ribosomal deoxyribonucleic acid (rDNA) of CJY/W1 had 99% sequence identity to that of Acanthamoeba sp. U/H-C1. Strains CJY/S1 and CJY/S2, isolated from soil, had similar mtDNA RFLP patterns, whereas strain CJY/W1, isolated from tap water, displayed a different pattern.
Conclusions
To the best of our knowledge, this is the first report on the identification of genotypes T4, T5, and T16 from environmental sources in Yanji, China.
Similar content being viewed by others
Background
Acanthamoeba species are widely distributed in the environment: their habitats include soil, freshwater, seawater, dust, and putrilage. Some Acanthamoeba species can cause keratitis, granulomatous amoebic encephalitis (GAE), pulmonary infections, cutaneous lesions, rhinosinusitis, osteomyelitis, or diffuse inflammation [1,2,3]. Acanthamoeba keratitis can lead to scarring of the cornea, resulting in a permanent visual impairment or complete blindness. GAE is a central nervous system disease that usually occurs in immunocompromised patients, such as those with acquired immune deficiency syndrome, systemic lupus erythematosus, and transplanted organ, or those undergoing chemotherapy for cancer. Nonetheless, several cases of GAE and skin infection due to Acanthamoeba spp. have been reported in immunocompetent individuals [4,5,6,7,8].
Eye infections have been reported to be caused by Acanthamoeba castellanii, Acanthamoeba polyphaga, Acanthamoeba rhysodes, Acanthamoeba culbertsoni, Acanthamoeba lugdunensis, Acanthamoeba griffini, Acanthamoeba hatchetti, Acanthamoeba quina, Acanthamoeba lenticulata, and Acanthamoeba triangularis. Central nervous system diseases have been caused by A. castellanii, A. culbertsoni, Acanthamoeba astronyxis, A. rhysodes, Acanthamoeba healyi, and A. lenticulata [6,10,11,12,, 9–13].
There have been few reports of Acanthamoeba spp. isolated from environmental samples in China [14]. In this study, we report the molecular biological characterization of four strains of Acanthamoeba (CJY/S1, CJY/S2, CJY/S3, and CJY/W1) isolated from soil and tap water in China. We identified these species by using mitochondrial DNA restriction fragment length polymorphism (mtDNA RFLP) analysis and 18S rDNA sequence alignment. We found that the three strains isolated from the soil belonged to the morphological group II and had genotypes T4 and T5, whereas the strain isolated from tap water belonged to the morphological group II and had genotype T16.
Methods
Isolation and cultivation of Acanthamoeba
Samples of soil and tap water were collected in Yanji, China. The samples were loaded onto 1.5% agar plates covered with heat-inactivated (60 °C for 1 h) Escherichia coli (American Type Culture Collection, ATCC 25922, free of plasmid). The plates were incubated at 25 °C, and growth of Acanthamoeba was observed under an inverted microscope on a daily basis for 1 week. Each cyst isolated with a glass capillary was inoculated on a new agar plate and incubated for 1 week. For axenization, a piece of agar (1 cm × 1 cm) covered with cysts was treated with 0.1 N HCl for 24 h, washed three times with sterile water, placed in peptone yeast glucose medium (10 g proteose peptone, 10 g yeast extract, 10 mL of 50% glucose, 10 mL of 0.5 M Na2HPO4, and 10 mL of 0.5 M K2HPO4 in 970 mL of sterile water), and incubated at 25 °C for 3 weeks. When most of the amoebae reached trophozoite stage, they were harvested and washed three times with phosphate-buffered saline.
Morphological examination
A cyst formed on the monoxenic plate was picked with a sterilized inoculating loop and transferred to a glass slide with a drop of sterile distilled water. The slide was then covered with a coverslip. Fifty cysts per plate were observed and measured under a Nomarski (differential interference contrast) microscope (Olympus, Japan).
Analysis of 18S rDNA sequences
Genomic DNA was extracted using phenol/chloroform method. The 18S rRNA gene was amplified using the following primers by Xuan et al. [13]: forward, 5′-CCGAATTCGTCGACAACCTGGTTGATCCTGCCAGT-3′; reverse, 5′-GGATCCAAGCTTGATCCTTCTGCAGGTTCACCTAC-3′. The amplified products were resolved by electrophoresis, recovered from the gel, and ligated into a T/A cloning vector (pGEM-T Easy Vector System I, Promega, USA) for subsequent transformation of E. coli. Positive clones were picked, and recombinant plasmid DNA was extracted using the Wizard® Plus Minipreps DNA Purification System (Promega, USA). Plasmids with inserts of the correct size were identified by EcoRI digestion and sequenced. The obtained final 18S ribosomal DNA (rDNA) sequences of the Acanthamoeba strains were deposited in GenBank (accession nos. KY827389–KY827392). The sequences were then compared with those of other Acanthamoeba sequences in GenBank using the Basic Local Alignment Search Tool (BLAST) search engine. Clustal X and GeneDoc were used for pairwise alignment and calculation of percent sequence dissimilarity. Phylogenetic analyses were performed, and the phylogenetic tree was drawn using the neighbor-joining (NJ) method with MEGA3 [15]. The reference strains of Acanthamoeba and the GenBank accession numbers of the 18S rDNA sequences used in this study are as follows: A. castellanii CDC:0981:V006 (T1), U07400; Acanthamoeba palestinensis (T2), U07411; A. griffini H37 (T3), S81337; A. castellanii Neff (T4), U07416; A. lenticulata E18-2 (T5), U94735; A. palestinensis 2802 (T6), AF019063; A. astronyxis R&H (T7), AF019064; Acanthamoeba tubiashi OC-15C (T8), AF019065; Acanthamoeba comandoni Pussard (T9), AF019066; A. culbertsoni A-1 (T10), AF019067; A. hatchetti BH-2 (T11), AF019068; A. healyi OC-3A (T12), AF019070; Acanthamoeba sp. UWC9 (T13), AF132134; Acanthamoeba sp. PN13 (T14), AF333609; Acanthamoeba jacobsi ATCC 30732 (T15), AY262360; Acanthamoeba sp. U/H-C1 (T16), AY026245; Acanthamoeba sp. E1a (T17), GU808277; Acanthamoeba byersi CDC:V621 (T18), KC822464; Acanthamoeba micheli BRO-2 (T19), KP711387; and Acanthamoeba sp. OSU 04-020 (T20), DQ451161.
Extraction of mtDNA and RFLP analysis
mtDNA of Acanthamoeba isolates was extracted using the method described by Yagita and Endo [16]. Briefly, amoebae were harvested and washed in cold phosphate-buffered saline, treated with TEG buffer (25 mM Tris–HCl, 10 mM ethylenediaminetetraacetic acid (EDTA), 50 mM glucose, pH 8.0), lysed with a fresh solution of 1% sodium dodecyl sulfate in 0.2 N NaOH and potassium acetate buffer, and left on ice for 30 min. mtDNA was then extracted with a phenol/chloroform mixture (1:1) and recovered by the precipitation with cold absolute ethanol in the presence of sodium acetate. The extracted mtDNA was then digested with EcoRI at 37 °C. The digested mtDNA was electrophoresed in 0.7% agarose gel and stained with ethidium bromide. The mtDNA RFLP patterns were observed and photographed.
Results
Morphology of Acanthamoeba isolates
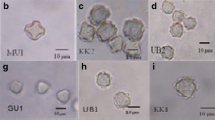
Cysts of the Acanthamoeba isolates CJY/S1, CJY/S2, CJY/S3, and CJY/W1 exhibited morphological characteristics typical of group II, as defined by Pussard and Pons [17]. They exhibited double-walled cyst morphology and featured thick, wrinkled ectocysts and satellite or polygonal endocysts (Fig. 1). The cyst diameter varied from 12.0 to 18.8 μm, and the number of arms was four to six (Table 1).
18S rRNA sequence analysis of the four isolated Acanthamoeba strains
The 18S rRNA genes of the Acanthamoeba sp. strains CJY/S1, CJY/S2, CJY/S3, and CJY/W1 were amplified using primers specific for Acanthamoeba spp. The polymerase chain reaction (PCR) products were cloned into the pGEM-T vector, and recombinant plasmids were digested with EcoRI. The full lengths of the 18S rRNA genes of the Acanthamoeba sp. strains CJY/S1, CJY/S2, CJY/S3, and CJY/W1 were 2255, 2252, 2292, and 2252 bp, respectively. Acanthamoeba sp. CJY/S1 and CJY/S2 had very high 18S rDNA sequence similarity with 18S rDNA sequences of A. castellanii Neff (99.82%) and Acanthamoeba sp. KA/E7 (99.69%). Acanthamoeba sp. CJY/S3 showed very high 18S rDNA sequence similarity with that of A. lenticulata 68–2 (99.74%). The 18S rRNA gene sequence of Acanthamoeba sp. CJY/W1 was closely related to the sequences of Acanthamoeba sp. U/H-C1 (99%) and Acanthamoeba sp. UWC9 (95%). Sequence alignment showed that 18S rDNA sequences of Acanthamoeba sp. strains CJY/S1 and CJY/S2 corresponded to genotype T4 and CJY/S3 corresponded to genotype T5, whereas the sequence of Acanthamoeba sp. CJY/W1 corresponded to genotype T16 (Fig. 2).
Dendrogram of Acanthamoeba strains based on 18S rDNA sequences. The neighbor-joining tree reflects the affiliation of the Acanthamoeba sp. strains CJY/S1, CJY/S2, CJY/S3, and CJY/W1 with the reference strains of genotypes T1–T20. T1: A. castellanii CDC: 0981:V006, GenBank accession no. U07400; T2: A. palestinensis, U07411; T3: A. griffini H37, S81337; T4: A. castellanii Neff, U07416; T5: A. lenticulata E18-2, U94735; T6: A. palestinensis 2802, AF019063; T7: A. astronyxis, AF019064; T8: A. tubiashi OC-15C, AF019065; T9: A. comandoni, AF019066; T10: A. culbertsoni A-1, AF019067; T11: A. hatchetti BH-2, AF019068; T12: A. healyi, AF019070; T13: Acanthamoeba sp. UWC9, AF132134; T14: Acanthamoeba sp. PN13, AF333609; T15: A. jacobsi ATCC 30732, AY262360; T16: Acanthamoeba sp. U/H-C1, AY026245; T17: Acanthamoeba sp. E1a, GU808277; T18: A. byersi CDC:V621, KC822464; T19: A micheli BRO-2, KP711387; T20: Acanthamoeba sp. OSU 04–020, DQ451161
mtDNA RFLP
Figure 3 shows agarose gel electrophoretic patterns of EcoRI-digested mtDNA extracted from the Acanthamoeba sp. strains CJY/S1, CJY/S2, CJY/S3, and CJY/W1. Acanthamoeba sp. strains CJY/S1 and CJY/S2 showed extremely similar mtDNA RFLP patterns, whereas Acanthamoeba sp. CJY/S3 and CJY/W1 each displayed a different pattern.
Discussion
There are many species of free-living amoebae. Some, such as Acanthamoeba spp., Naegleria spp., Hartmannella spp., or Balamuthia mandrillaris, are opportunists that can cause infections in humans and animals [18, 19]. Naegleria and Acanthamoeba species have been identified as causes of serious human infections. Some species of Acanthamoeba can cause amoebic keratitis, particularly in contact lens wearers and immunocompromised individuals experiencing subacute or chronic central nervous system infections [20, 21].
Eighteen species of Acanthamoeba have been classified into three groups according to the shape and size of cysts. Species in group I are nonpathogenic except A. astronyxis, A. byersi and A. comandoni [22, 23]. Most of the pathogenic Acanthamoeba species belong to group II. Species in group III, such as A. culbertsoni, A. healyi, and A. lenticulata, often cause infections of the brain. The cyst morphology of the four isolates characterized in this study resembled that of various species within morphological group II. However, the classification of Acanthamoeba spp. based on morphological characteristics has proven to be unreliable. The morphology of Acanthamoeba spp. may change depending on culture conditions. Furthermore, different Acanthamoeba species in the same group can have similar morphology, and Acanthamoeba cysts of two species may show only transient differences, thereby causing difficulties in the identification of the species.
Lass et al. reported partial sequences of T4 strains in environmental samples in China recently [14], which is different from our data which is full of 18S sequences of four distinct genotypes of isolated strains. At present, sequence analysis of genomic DNA is considered the method of choice for identifying species of Acanthamoeba. Sequence analysis of 18S rRNA genes is frequently used.
Based on the nucleotide sequence of 18S rDNA, Acanthamoeba was initially clustered into 12 genotypes, from T1 to T12 [12, 24]. Recently, new genotypes of Acanthamoeba have been identified [25,26,27,28,29]. Five genotypes of Acanthamoeba are associated with keratitis: T4 is the primary genotype and T3 is the secondary genotype, whereas Acanthamoeba species of the remaining genotypes (T5, T6, and T2) are considered to be rare causes of this disease. Gast [28] reported that sequence differences among 15 strains of Acanthamoeba within genotype T4 were in the range of 0–4%, whereas sequence differences among genotypes were 6–12%.
In this study, the full-length 18S rRNA genes from the Acanthamoeba sp. strains CJY/S1, CJY/S2, CJY/S3, and CJY/W1, isolated from soil and tap water of Yanji, China, were determined to be 2255, 2252, 2292, and 2252 bp, respectively. These lengths are close to the range of 2300–2700 bp reported by Stothard et al. [12]. The 18S rDNA sequences of the Acanthamoeba sp. strains CJY/S1, CJY/S2, CJY/S3, and CJY/W1 were compared with those of reference strains of genotypes T1–T20, which were obtained from GenBank by using BLAST searches. Pairwise alignment and calculation of the percent sequence dissimilarity using Clustal X and GeneDoc showed that Acanthamoeba sp. strains CJY/S1 and CJY/S2 had genotype T4, which encompasses the majority of clinical and environmental isolates of Acanthamoeba. In a phylogenetic tree based on the 18S rDNA sequence, Acanthamoeba CJY/S3 strain was positioned close to genotype T5 species and was related to A. lenticulata. The Acanthamoeba sp. strains CJY/S1 and CJY/S2 showed extremely similar mtDNA RFLP patterns. Acanthamoeba sp. CJY/W1 had genotype T16 and was closely related to Acanthamoeba sp. U/H-C1 (99%) and Acanthamoeba sp. UWC9 (95%) [30]. Acanthamoeba sp. CJY/W1, isolated from tap water, had a mtDNA RFLP pattern different from those of the Acanthamoeba sp. strains CJY/S1, CJY/S2, and CJY/S3, which were isolated from soil.
Recent studies have shown that A. castellanii (genotype T4) and A. lenticulata (genotype T5) can infect the cornea and central nervous system in humans, but there are insufficient reports pertaining to strains of genotype T16. Further research is required to determine whether the Acanthamoeba sp. strains CJY/S1, CJY/S2, CJY/S3, and CJY/W1 isolated by us from environmental samples could be pathogenic to humans and animals.
Conclusions
Strains CJY/S1 and CJY/S2, isolated from soil, had similar mtDNA RFLP patterns, whereas strain CJY/W1, isolated from tap water, displayed a different pattern. To the best of our knowledge, this is the first report on the identification of genotypes T4, T5, and T16 from environmental sources in Yanji, China.
Abbreviations
- ATCC:
-
American Type Culture Collection
- BLAST:
-
Basic Local Alignment Search Tool
- EDTA:
-
Ethylene Diamine Tetraacetic Acid
- GAE:
-
Granulomatous amoebic encephalitis
- mtDNA RFLP:
-
Mitochondrial DNA restriction fragment length polymorphism
- PCR:
-
Polymerase chain reaction
- rDNA:
-
ribosomal deoxyribonucleic acid
References
Lorenzo-Morales J, Khan NA, Walochnik J. An update on Acanthamoeba keratitis: diagnosis, pathogenesis and treatment. Parasite. 2015;22:10.
Morrison AO, Morris R, Shannon A, Lauer SR, Guarner J, Kraft CS. Disseminated Acanthamoeba infection presenting with cutaneous lesions in an immunocompromised patient: a case report, review of histomorphologic findings, and potential diagnostic pitfalls. Am J Clin Pathol. 2016;145:266–70.
Steinberg JP, Galindo RL, Kraus ES, Ghanem KG. Disseminated acanthamebiasis in a renal transplant recipient with osteomyelitis and cutaneous lesions: case report and literature review. Clin Infect Dis. 2002;35:e43–9.
Castillo RD, Garza JX, Shamszadeh M, Reiff AO, Marzan KA. Acanthamoeba meningoencephalitis presenting as neuropsychiatric lupus in a pediatric patient. Clin Exp Rheumatol. 2012;30:272–6.
Galarza C, Ramos W, Gutierrez EL, Ronceros G, Teran M, Uribe M, et al. Cutaneous acanthamebiasis infection in immunocompetent and immunocompromised patients. Int J Dermatol. 2009;48:1324–9.
Lackner P, Beer R, Broessner G, Helbok R, Pfausler B, Brenneis C, et al. Acute granulomatous acanthamoeba encephalitis in an immunocompetent patient. Neurocrit Care. 2010;12:91–4.
Martinez-Giron R, Esteban JG, Ribas A, Doganci L. Protozoa in respiratory pathology: a review. Eur Respir J. 2008;32:1354–70.
Nachega JB, Rombaux P, Weynand B, Thomas G, Zech F. Successful treatment of Acanthamoeba rhinosinusitis in a patient with AIDS. AIDS Patient Care STDs. 2005;19:621–5.
Iovieno A, Oechsler RA, Ledee DR, Miller D, Alfonso EC. Drug-resistant severe Acanthamoeba keratitis caused by rare T5 Acanthamoeba genotype. Eye Contact Lens. 2010;36:183–4.
Polat ZA, Obwaller A, Vural A, Walochnik J. Efficacy of miltefosine for topical treatment of Acanthamoeba keratitis in Syrian hamsters. Parasitol Res. 2012;110:515–20.
Schaumberg DA, Snow KK, Dana MR. The epidemic of Acanthamoeba keratitis: where do we stand? Cornea. 1998;17:3–10.
Stothard DR, Schroeder-Diedrich JM, Awwad MH, Gast RJ, Ledee DR, Rodriguez-Zaragoza S, et al. The evolutionary history of the genus Acanthamoeba and the identification of eight new 18S rRNA gene sequence types. J Eukaryot Microbiol. 1998;45:45–54.
Xuan YH, Chung BS, Hong YC, Kong HH, Hahn TW, Chung DI. Keratitis by Acanthamoeba triangularis: report of cases and characterization of isolates. Korean J Parasitol. 2008;46:157–64.
Lass A, Guerrero M, Li X, Karanis G, Ma L, Karanis P. Detection of Acanthamoeba spp. in water samples collected from natural water reservoirs, sewages, and pharmaceutical factory drains using LAMP and PCR in China. Sc Total Environ. 2017;584–585:489–94.
Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–63.
Yagita K, Endo T. Restriction enzyme analysis of mitochondrial DNA of Acanthamoeba strains in Japan. J Protozool. 1990;37:570–5.
Pussard M, Pons R. Morphologie de la paroi kystique et taxonomie du gene Acanthamoeba (Protozoa, Amoebida). Protistologica. 1977;13:557–98.
Trabelsi H, Dendana F, Sellami A, Sellami H, Cheikhrouhou F, Neji S, et al. Pathogenic free-living amoebae: epidemiology and clinical review. Pathol Biol. 2012;60:399–405.
Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Microbiol. 2007;50:1–26.
Alkhunaizi AM, Dawamneh MF, Banda RW, Daabil RA, Al-Tawfiq JA, Akkad SA, et al. Acanthamoeba encephalitis in a patient with systemic lupus treated with rituximab. Diagn Microbiol Infect Dis. 2013;75:192–4.
Sarica FB, Tufan K, Cekinmez M, Erdogan B, Altinors MN. A rare but fatal case of granulomatous amebic encephalitis with brain abscess: the first case reported from Turkey. Turk Neurosurg. 2009;19:256–9.
Hajialilo E, Behnia M, Tarighi F, Niyyati M, Rezaeian M. Isolation and genotyping of Acanthamoeba strains (T4, T9, and T11) from amoebic keratitis patients in Iran. Parasitol Res. 2016;115:3147–51.
Qvarnstrom Y, Nerad TA, Visvesvara GS. Characterization of a new pathogenic Acanthamoeba species, A. byersi n. sp., isolated from a human with fatal amoebic encephalitis. J Eukaryot Microbiol. 2013;60:626–33.
Gast RJ, Ledee DR, Fuerst PA, Byers TJ. Subgenus systematics of Acanthamoeba: four nuclear 18S rDNA sequence types. J Eukaryot Microbiol. 1996;43:498–504.
Corsaro D, Venditti D. Phylogenetic evidence for a new genotype of Acanthamoeba (Amoebozoa, Acanthamoebida). Parasitol Res. 2010;107:233–8.
Corsaro D, Walochnik J, Kohsler M, Rott MB. Acanthamoeba misidentification and multiple labels: redefining genotypes T16, T19, and T20 and proposal for Acanthamoeba micheli sp. nov. (genotype T19). Parasitol Res. 2015;114:2481–90.
Di Cave D, Monno R, Bottalico P, Guerriero S, D’Amelio S, D’Orazi C, et al. Acanthamoeba T4 and T15 genotypes associated with keratitis infections in Italy. Eur J Clin Microbiol Infect Dis. 2009;28:607–12.
Gast RJ. Development of an Acanthamoeba-specific reverse dot-blot and the discovery of a new ribotype. J Eukaryot Microbiol. 2001;48:609–15.
Nuprasert W, Putaporntip C, Pariyakanok L, Jongwutiwes S. Identification of a novel T17 genotype of Acanthamoeba from environmental isolates and T10 genotype causing keratitis in Thailand. J Clin Microbiol. 2010;48:4636–40.
Horn M, Fritsche TR, Gautom RK, Schleifer KH, Wagner M. Novel bacterial endosymbionts of Acanthamoeba spp. related to the Paramecium caudatum symbiont Caedibacter caryophilus. Environ Microbiol. 1999;1:357–67.
Acknowledgements
Not applicable.
Funding
This work is supported by the Public Health Research Center at Jiangnan University (No. JUPH201501) and Jiangsu Science and Technology Department (BM2015024-3).
Availability of data and materials
Please contact the author for data requests.
Authors’ contributions
YHX carried out the molecular genetic studies and drafted the manuscript. YQS participated in the design of the study and carried out the molecular genetic studies. GY carried out the mtDNA RFLP. YXG participated in the sequence alignment. SZZ conceived of the study, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Xuan, Y., Shen, Y., Ge, Y. et al. Isolation and identification of Acanthamoeba strains from soil and tap water in Yanji, China. Environ Health Prev Med 22, 58 (2017). https://doi.org/10.1186/s12199-017-0655-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12199-017-0655-2