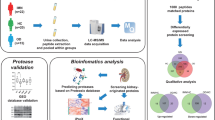
Abstract
Background
Primary membranous nephropathy (PMN) is an important cause of nephrotic syndrome in adults. Urine proteome may provide important clues of pathophysiological mechanisms in PMN. In the current study, we analyzed and compared the proteome of urine from patients with PMN and normal controls.
Methods
We performed two technical replicates (TMT1 and TMT2) to analyze and compare the urine proteome from patients with PMN and normal controls by tandem mass tag (TMT) technology coupled with nanoscale liquid chromatography tandem mass spectrometry analysis (LC–MS/MS). Gene ontology (GO) enrichment analysis was performed to analyse general characterization of the proteins. The proteins were also matched against the database of Kyoto Encyclopedia of Genes and Genomes (KEGG). For validation, Western blot was used to analyze the selected proteins.
Results
A total of 509 proteins and 411 proteins were identified in TMT1 and TMT2, respectively. 249 proteins were both identified in two technical replicates. GO analysis and KEGG analysis revealed immunization and coagulation were predominantly involved. Among the differential protein, the overexcretion of alpha-1-antitrypsin (A1AT) and afamin (AFM) were validated by Western blot analysis.
Conclusions
Our data showed the important role of immunologic mechanism in the development of PMN, and the value of urinary A1AT and AFM in biomarker discovery of patients with PMN. The discovery of the overexcretion of A1AT and AFM in the urine can help to further elucidate pathogenetic mechanisms involved in PMN.
Similar content being viewed by others
Background
Primary membranous nephropathy (PMN) is the most common cause of nephrotic syndrome in the adult population [1, 2] and it is one of the most common diseases affecting the glomerulus [2]. The kidney filtration barrier consists of capillary endothelial cells, glomerular basement membrane and highly specialized epithelial cell, the podocytes [3, 4]. In patients with PMN, this filtration barrier is injured, leading to massive loss of proteins in urine (proteinuria) [5]. Histopathologically, this disorder is characterized by discovery of immune complexes deposits in a specific part of glomerulus between the glomerular basement membrane and podocyte (called subepithelium), which contain podocyte antigens or planted antigens and circulating antibodies specific to those antigens, resulting in complement activation [2]. This discovery corroborates the hypothesis that the most likely pathogenesis of PMN is autoimmune.
Tremendous insights into the pathophysiology of PMN have been made recently, with studies that have identified M-type phospholipase A2 receptor (PLA2R) [6] and thrombospondin type-1 domain-containing 7A (THSD7A) [7] as two major autoantigens in PMN. These finding supported the theory that podocytes act as sources of antigens for the formation of subepithelial immune complexes deposits [8]. Serum anti-PLA2R antibody identifies approximately 60–80% of cases of PMN [9,10,11]. Despite the undeniable potential of anti-PLA2R antibody as a biomarker of disease in patients with PMN, this antibody does not explain the etiology of the disease in a substantial proportion of cases [12].
Despite progress in the understanding of pathogenesis, the diagnosis of PMN is definitively dependent on only renal biopsy [13]. The exact pathogenesis of PMN remains unknown, but podocytes and podocytes-related proteins appear to have a pivotal role in the development of PMN. The pathogenicity of anti-PLA2R has not yet been confirmed [14]. However, human anti-THSD7A has recently been shown to induce PMN with proteinuria in mice [15]. The morphology of healthy podocyte foot processes is necessary for maintaining the characteristics of the kidney filtration barrier [16]. Interaction of circulating autoantibodies with antigens at the podocyte cell membrane-basement membrane interface generally is regarded as the fundamental pathological mechanism [17]. Since the visceral epithelial cell of Bowman’s capsule is podocytes, the content of podocytes will be released into urine when podocytes were destroyed by membrane attack complex. Therefore, urine can be considered as a potential source to provide important clues of pathophysiological mechanisms of PMN.
Nanoscale liquid chromatography tandem mass spectrometry analysis (LC–MS/MS) may be considered a novel method for identifying candidate biomarkers for patients suffering from PMN [18]. Tandem mass tag (TMT) is a label-based quantification technology that enables accurate and simultaneous comparison of multiple samples for protein and peptide quantification [19]. TMT coupled with LC–MS/MS has the ability to analyze hundreds of proteins with the high-resolution, mass accuracy, and sensitivity [20]. LC–MS/MS has been applied in the proteomic analysis of various kidney diseases, such as acute kidney injury [21], lupus nephritis [22], diabetic nephropathy [23] and IgA nephropathy [24]. However, proteomic analysis of PMN has been rarely studied, especially in urine.
Our aim was to test urine proteomics as a non-invasive method for identification of new protein biomarkers of PMN in urine, and link them to pathogenesis of the disease through known signaling and metabolic pathways. In this study, we performed proteomic analysis using TMT technology coupled with LC–MS/MS, comparing PMN urine samples with normal control groups. The proteomic results were validated with Western blot, providing the preliminary evidence of protein biomarkers in urine of PMN patients.
Methods
Sample collection
Midstream morning urine samples were collected from patients of biopsy proven PMN with positive anti-PLA2R antibody (Group A, n = 32) and negative anti-PLA2R antibody (Group B, n = 31). Patient characteristics are shown in Additional file 1: Table S1 and Additional file 2: Table S2. Cases of secondary MN were excluded from the present study, in particular patients with systemic autoimmunity diseases, viral hepatitis B and C and HIV infection, neoplastic conditions and exposure to toxic agents. All samples were collected at the time of biopsy. Urine samples from healthy volunteers (Group C, n = 32) were also collected (Additional file 3: Table S3). Samples were kept for less than 4 h at room temperature followed by low speed centrifugation at 2000×g, 10 min at room temperature to remove cellular debris and then stored at − 80 °C until use.
At first, we performed the first TMT experiment (called TMT1 in this study). To increase accuracy repeatability, we performed another TMT experiment (called TMT2 in this study) by increasing the sample size and replicates to validate TMT1 (Table 1). Samples from group A, group B and group C (9 patients in each group) were tested by Western blot (Additional file 1: Table S1, Additional file 2: Table S2, Additional file 3: Table S3).
This study was approved by the ethics committee of Peking University First Hospital and informed consents were obtained from all participants.
Protein precipitation
The 10 mL sample aliquots from each participant were thawed, and 2.5 mL of trichloroacetic acid (TCA) precipitation solution (30% v/v) were added to the sample aliquots to a final TCA concentration of 6% v/v. Strong vortexing for 1 min and overnight incubation at − 20 °C were performed. The mixtures were centrifuged at 14,000×g for 15 min at 4 °C and the supernatants were discarded. In order to eliminate traces of acid that can negatively affect the digestion efficiency, the pellets were resuspended in 2.5 mL of chilled acetone (− 20 °C) and clarified by centrifugation at 14,000×g at 25 °C for 10 min. The wash step was repeated once more. The supernatants were discarded and the pellets were dried naturally. The dry pellets were resuspended in 300 μL solubilization buffer (8 mol/L urea and 0.1 mol/L ammonium bicarbonate), vortexed strongly for 1 min and incubated for 15 min at 37 °C. Protein concentration was determined using the bicinchoninic acid protein assay.
Protein preparation
We pooled the samples from each participant on equal quantity to form three pooled samples in TMT1 and six pooled samples in TMT2 (Table 1). 200 μL reducing solution [10 mg/mL dithiothreitol (DTT)] was added and incubate for 1 h at 37 °C. Then, 200 μL alkylation solution [12 mg/mL iodoacetamide (IAA)] was added to block reduction of cysteine residues and incubated for 1 h in the dark. The DTT, IAA and other low-molecular-weight components were removed using trypsin buffer (1 mmol/L CaCl2 and 100 mmol/L Tris–HCl, pH 8.0) by repeated ultrafiltration (3 kD Microcon; Millipore Corp., Billerica, MA, USA). The samples were then digested with trypsin with the ratio of protein:trypsin = 100:1 at 37 °C overnight.
TMT labeling
Protein peptides (50 μg) from each group were processed strictly according to the manufacturer’s protocol for TMT Mass Tag Labeling Kits and Reagents (Thermo Fisher Scientific, Waltham, MA, USA). The protein peptides were labeled randomly with 126, 130, 131 TMTsixplex tags for TMT1 and 126, 127C, 127N, 128N, 129C, 130C TMT10plex for TMT2 (Table 1). The TMT Label Reagents were equilibrated to room temperature and reconstituted with 41 μL of anhydrous acetonitrile. The reagents were dissolved for 5 min with occasional vortexing and then centrifuged to gather the solution. The TMT Label Reagents were added to the corresponding peptide samples and incubated the reaction for 1 h at room temperature. 8 μL of 5% hydroxylamine were added to the sample and incubate for 15 min to quench the reaction. Samples from three groups of TMT1 and six groups of TMT2 were mixed equally and lyophilized, respectively. 1 μL aliquot of sample was removed from each group to test labeling and extraction efficiency, and the sample was subjected to a matrix assisted laser desorption ionization procedure after Ziptip desalting.
Strong cationic exchange chromatography
The mixed peptides were dissolved in buffer A (2% acetonitrile (ACN) and 20 mmol/L ammonium formate, pH 10.0). Then, the samples were loaded onto a reverse-phase column (Luna C18, 4.6 × 150 mm; Phenomenex, Torrance, CA, USA) and eluted using a step linear elution program: 0–10% buffer B (500 mmol/L KCl, 10 mmol/L KH2PO4 in 25% ACN, pH 2.7) for 10 min, 10–20% buffer B for 25 min, 20–45% buffer B for 5 min and 50–100% buffer B for 5 min at a flow rate of 0.7 mL/min. The samples were collected each min and centrifuged for 5–45 min. The fractions (about 40) collected were finally combined into 10 pools and desalted on C18 Cartridges (Empore™ standard density SPE C18 Cartridges, bed I.D. 7 mm, 3 mL volume; Sigma, St. Louis, MO, USA).
LC–MS/MS analysis
The reconstituted peptides were analyzed with the Q-Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) coupled with a nano high-performance liquid chromatography (UltiMate 3000 LC Dionex; Thermo Fisher Scientific, Waltham, MA, USA) system. The peptides were loaded onto a C18 reversed phase column (3 μm C18 resin, 75 μm × 15 cm) and separated on an analytical column (5 μm C18 resin, 150 μm × 2 cm; Dr. Maisch GmbH, Ammerbuch, Germany) using mobile phase A: 0.5% formic acid (FA)/H2O and B: 0.5% FA/ACN at a flow rate of 300 nL/min, using a 150 min gradient. Spectra were acquired in data-dependent mode. The 10 most intense ions were selected for MS scanning (300–1800 m/z, 60,000 resolution at 400 m/z, accumulation of 1 × 106 ions for a maximum of 500 ms, 1 microscan). The isolation window was 1.3 m/z and the MS/MS spectra were accumulated for 150 ms using an Orbitrap. MS/MS spectra were measured at resolution of 15,000 at 400 m/z. Dynamic precursor exclusion was allowed for 2 min after each MS/MS spectrum measurement and was set to 17,500 at 200 m/z. Normalized collision energy was 30 eV and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at the maximum fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled. All of the MS proteomics data have been deposited to the integrated proteome resources [25] with the accession number IPX0001113000.
Sequence database search and data analysis
The raw mass data are processed for the peptide data analysis using Proteome Discoverer (v 1.4.0.288; Thermo Fisher Scientific, Waltham, MA, USA) with a false discovery rate (FDR) < 1% and expected cutoff or ion score < 0.05 (with 95% confidence) for searching the UniProt Human Complete Proteome database: digestion with trypsin with no more than two missed cleavages, TMT modification to lysine side chains and N-termini, and variable oxidation of methionine residues. A mass tolerance of 10 ppm for intact peptide masses and 0.6 Da for fragmented ions were permitted. All peptide spectra scoring higher against a reversed human sequence were eliminated from further consideration. Protein probabilities were assigned using the Protein Prophet algorithm and proteins with at least one unique peptide were identified. The upregulated or downregulated proteins in three replicates with relative quantification 1.5 fold-changes were selected as being differentially expressed in the data.
Bioinformatics analyses
We performed gene ontology (GO) bioinformatics analysis on the differentially expressed proteins with a 1.5-fold change to catalog the molecular functions, cellular components and biological processes [26]. The enrichment analysis of the proteins was conducted with all proteins GO biological processes database information from our study using the following formula [27]:
where N is the number of all proteins within the GO annotation information, n is the number of differentially regulated proteins within GO annotation information, M is the number of proteins for a given GO annotation, and m is the number of differentially regulated proteins following a given GO annotation. Significant GO enrichment was defined as P ≤ 0.05.
The interactions among these proteins regarding the biological pathways were determined using Kyoto Encyclopedia of Genes and Genomes (KEGG) database to better understand these differentially proteins in relation to the published literature. The Pathway Maps tool was used to enrich the pathways with all proteins KEGG database information from our study and P values were calculated based on a hypergeometric distribution. Significant pathway enrichment was defined as P ≤ 0.05.
Western blot analysis
The increases of alpha-1-antitrypsin (A1AT) and afamin (AFM) were validated by Western blot. For internal validation, due to the inadequate protein, we only validated with 13 samples of Group A, 13 samples of Group B and 10 samples of Group C. For external validation, nine samples for each group were validated. The datail characteristics of the samples are shown in Additional file 1: Table S1, Additional file 2: Table S2 and Additional file 3: Table S3. The protein samples were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred onto polyvinylidene fluoride membranes and blocked with 5% nonfat milk at room temperature for 1 h. Mouse monoclonal antibody against A1AT (1:1000, ab9399; Abcam, Cambridge, MA, USA) and mouse monoclonal antibody against AFM (1:500, sc-373849; Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used as primary antibody. Goat anti-mouse horse radish peroxidase (HRP)-conjugated IgG (1:5000, sc-2005; Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used as secondary antibody. Signals were developed using Immobilon Western HRP substrate (WBKLS0100; Millipore, Billerica, MA, USA).
Statistical analysis
Statistical analyses were performed using the SPSS software version 19.0 for Windows (IBM, Chicago, IL, USA). Graphs were prepared using GraphPad Prism version 6.0 (GraphPad Software, San Diego, CA, USA). Values are represented as means ± standard deviations. Student t test was used to compare differences between continuous data. A two-tailed P value < 0.05 was considered to be statistically significant.
Results
GO analysis and KEGG analysis showed immunization and coagulation were predominantly involved.
A total of 509 proteins and 411 proteins were identified in TMT1 and TMT2, respectively (Additional file 4: Table S4, Additional file 5: Table S5). 249 proteins were identified in both TMT1 and TMT2.
We performed GO analysis to enrich and cluster the 249 differential proteins. It was revealed that most of the proteins were involved in the platelet degranulation (n = 31), complement activation by classical pathway (n = 27), innate immune response (n = 26), cell adhesion (n = 26), receptor-mediated endocytosis (n = 26) and immune response (n = 25) (Fig. 1). GO enrichment analysis of biological processes showed that platelet degranulation (P < 0.05) is the main GO term associated with these proteins (Fig. 2). The detail information of GO enrichment of molecular functions, cellular components and biological processes was shown in Additional file 6: Table S6.
In addition, GO annotation analysis revealed that the subcellular proteins were distributed in the extracellular exosome (n = 224), extracellular space (n = 132), extracellular region (n = 126) and plasma membrane (n = 94) (Fig. 3) and associated with calcium ion binding (n = 35), serine-type endopeptidase activity (n = 32), antigen binding (n = 19) and identical protein binding (n = 18) (Fig. 4).
The KEGG pathway annotation analysis indicated that complement and coagulation cascades (n = 20, P < 0.05) is the main KEGG pathway associated with these proteins (Figs. 5 and 6). KEGG function classification analysis revealed that immune system (n = 36) and signaling molecules and interaction (n = 27) were predominantly involved (Fig. 7). The detail information of KEGG pathway enrichment was shown in Additional file 7: Table S7.
AFM and A1AT were increased in urine from patients with PMN
Intersecting the set of differential proteins in the TMT1 and TMT2, 32 proteins and 38 proteins were found to be upregulated based on the 1.5 fold-changes of group A/C and group B/C respectively. The upregulated proteins were highly consistent in both groups, resulting in 30 proteins identified by both group A/C and group B/C (Fig. 8). The top 10 proteins of mean fold change were listed in Table 2. The maximum differentially abundant protein enriched in both groups was alpha-1-antitrypsin (A1AT), a serine proteinase inhibitor. In addition, we selected afamin from the top 10 proteins for further analysis.
The increase of A1AT and AFM were validated by Western blot
The findings of the proteomic analysis were validated by Western blot. As shown in Fig. 9, A1AT and AFM were significantly increased in PMN urine compared to control samples.
Overexcretion of alpha-1-antitrypsin (A1AT) and afamin (AFM) in urine from patients with PMN. a Representative images of Western blot. Bar chats of relative fold-change of A1AT (b) and AFM (c) of internal validation, and A1AT (d) and AFM (e) of external validation. Asterisk denotes P < 0.05 using student t test. The bars represent the means ± standard deviations
Discussion
Urine proteome may provide important clues of pathophysiological mechanisms in PMN because urine samples are kidney-derived and easily acquirable. The expression of lysosome membrane protein-2 in urinary microvesicles has been suggested to be a biomarker for PMN [28], but this study only focused on urinary microvesicles. When podocytes, one of the layers of filtration barrier, were destroyed through the way of complement mediated cytolysis, the cell lysate will be released into urine. In the present study, we demonstrate the value of urine in biomarker discovery in PMN. Comparison of the urinary proteome of patients with PMN to normal controls enabled identification of new proteins possibly involved in the pathogenesis of PMN. This pilot study confirmed that the application of our approach is successful in identifying proteins upregulated in PMN and found that A1AT and AFM were significantly increased in PMN urine compared to control samples.
By GO function analysis and KEGG pathway analysis, our study demonstrated immunization was predominantly involved, including complement activation and immune response found in biological process analysis and complement cascades and immune system found in pathway analysis. It has been clarified PMN is driven by an underlying immune response [2]. The immune complexes cause podocyte injury characterized by actin–cytoskeleton disorganization and podocyte foot-process effacement, leading to increased permeability of the glomerular filtration barrier and eventually to massive proteinuria and nephrotic syndrome [29]. In addition, our study discovered that coagulation was also involved, including platelet degranulation found in biological process analysis and coagulation cascades found in pathway analysis. PMN is the pathological type of nephrotic syndrome associated with the highest incidence of thromboembolic events [30]. The underlying mechanisms of PMN-related hypercoagulability are not fully understood [31] and greater efforts should be made to clarify whether platelet degranulation was involved in PMN-related thromboembolic events.
A1AT, also named α1-Pi (α1 proteinase inhibitor) and SERPINA1 (serine protease inhibitor, group A, member 1), is the most abundant serum serine protease inhibitor [32]. Its main physiological role is to inhibit the activity of different endogenous serine proteases, such as elastase, myeloperoxidase and proteinase-3 [32, 33]. Due to its anti-protease activity, A1AT can exert tissue-protective effects. A1AT levels are upregulated in the renal tissue of adenine-induced chronic renal failure model [34] and in vivo administration of clinical grade A1AT improves renal function, decreases acute tubular necrosis and ameliorates acute kidney injury following experimental kidney ischemia reperfusion damage [35]. The overexcretion of A1AT have been described in the urine of IgA nephropathy, membranoproliferative glomerulonephritis, minimal change disease, focal segmental glomerulosclerosis and membranous nephropathy [36, 37]. Upregulation of A1AT could lead to inhibition of neutrophil elastase, which can contribute to accumulation of mesangial matrix, maintaining the elasticity of blood vessels and glomerular integrity [34]. A1AT is located in the cytoplasm of podocytes within sclerotic glomeruli [13] and its relationship with podocyte dysfunction and podocyte stress need to be further investigated.
AFM is a glycoprotein that is present in biological fluids such as plasma, urine, cerebrospinal, ovarian follicular and seminal fluids [38]. It is the fourth member of the human albumin gene family, which includes albumin, α-fetoprotein and vitamin D-binding protein [39]. Comparative proteomics have identified AFM as a potential urine biomarker for focal segmental glomerulosclerosis [40], paediatric idiopathic nephrotic syndrome [41] and IgA nephropathy [42]. Further studies in larger patient population are needed to investigate the role of AFM in kidney diseases.
This study has several limitations. To increase accuracy and reduce variability, we performed TMT2 by increasing the sample size and replicates to validate TMT1, resulting in the mismatch of of biological samples added into the pool. We did not assess other clinically pathological pattern of glomerular diseases, thus we did not clearly confirm the effect of differential diagnosis of A1AT and AFM. In addition, we only validated the candidates proteins in a small sized sample set by Western blot, clinical usefulness need to be verified with large sample size.
Conclusions
In conclusion, we succeeded in performing urine proteomics of PMN using TMT technology coupled with LC–MS/MS. According to the GO and KEGG analyses, it was demonstrated that these proteins are involved in multiple biological processes and pathways, but mainly in immune response and coagulation cascades. Interestingly, A1AT and AFM exhibited significantly changes and verified by Western blot. Further research is needed to explore the role of these protein as the candidate biomarkers of PMN and functions in pathogenesis.
Abbreviations
- PMN:
-
primary membranous nephropathy
- GO:
-
gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- A1AT:
-
alpha-1-antitrypsin
- AFM:
-
afamin
- PLA2R:
-
M-type phospholipase A2 receptor
- THSD7A:
-
thrombospondin type-1 domain-containing 7A
- TMT:
-
tandem mass tag
- LC–MS/MS:
-
liquid chromatography coupled with tandem mass spectrometry
- TCA:
-
trichloroacetic acid
- DTT:
-
dithiothreitol
- IAA:
-
iodoacetamide
- ACN:
-
acetonitrile
- FA:
-
formic acid
- HRP:
-
horse radish peroxidase
- SH2:
-
src homology 2
- SH3:
-
src homology 3
- α1-Pi:
-
α1 proteinase inhibitor
- SERPINA1:
-
serine protease inhibitor, group A, member 1
References
McGrogan A, Franssen CF, de Vries CS. The incidence of primary glomerulonephritis worldwide: a systematic review of the literature. Nephrol Dial Transplant. 2011;26:414–30.
Ronco P, Debiec H. Pathophysiological advances in membranous nephropathy: time for a shift in patient’s care. Lancet. 2015;385:1983–92.
Grahammer F. New structural insights into podocyte biology. Cell Tissue Res. 2017. https://doi.org/10.1007/s00441-017-2590-3.
Brahler S, Yu H, Suleiman H, Krishnan GM, Saunders BT, Kopp JB, et al. Intravital and kidney slice imaging of podocyte membrane dynamics. J Am Soc Nephrol. 2016;27:3285–90.
Sinico RA, Mezzina N, Trezzi B, Ghiggeri GM, Radice A. Immunology of membranous nephropathy: from animal models to humans. Clin Exp Immunol. 2016;183:157–65.
Beck LH Jr, Bonegio RG, Lambeau G, Beck DM, Powell DW, Cummins TD, et al. M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N Engl J Med. 2009;361:11–21.
Tomas NM, Beck LH Jr, Meyer-Schwesinger C, Seitz-Polski B, Ma H, Zahner G, et al. Thrombospondin type-1 domain-containing 7A in idiopathic membranous nephropathy. N Engl J Med. 2014;371:2277–87.
Francis JM, Beck LH Jr, Salant DJ. Membranous nephropathy: a journey from bench to bedside. Am J Kidney Dis. 2016;68:138–47.
Pang L, Zhang AM, Li HX, Du JL, Jiao LL, Duan N, et al. Serum anti-PLA2R antibody and glomerular PLA2R deposition in Chinese patients with membranous nephropathy: a cross-sectional study. Medicine (Baltimore). 2017;96:e7218.
Pourcine F, Dahan K, Mihout F, Cachanado M, Brocheriou I, Debiec H, et al. Prognostic value of PLA2R autoimmunity detected by measurement of anti-PLA2R antibodies combined with detection of PLA2R antigen in membranous nephropathy: a single-centre study over 14 years. PLoS ONE. 2017;12:e0173201.
Obrisca B, Ismail G, Jurubita R, Baston C, Andronesi A, Mircescu G. Antiphospholipase A2 receptor autoantibodies: a step forward in the management of primary membranous nephropathy. Biomed Res Int. 2015;2015:249740.
Cattran DC, Brenchley PE. Membranous nephropathy: integrating basic science into improved clinical management. Kidney Int. 2017;91:566–74.
Smith A, L’Imperio V, De Sio G, Ferrario F, Scalia C, Dell’Antonio G, et al. Alpha-1-Antitrypsin detected by MALDI imaging in the study of glomerulonephritis: its relevance in chronic kidney disease progression. Proteomics. 2016;16:1759–66.
Couser WG. Primary membranous nephropathy. Clin J Am Soc Nephrol. 2017;12:983–97.
Tomas NM, Hoxha E, Reinicke AT, Fester L, Helmchen U, Gerth J, et al. Autoantibodies against thrombospondin type 1 domain-containing 7A induce membranous nephropathy. J Clin Investig. 2016;126:2519–32.
Scott RP, Quaggin SE. Review series: the cell biology of renal filtration. J Cell Biol. 2015;209:199–210.
Glassock RJ. The pathogenesis of idiopathic membranous nephropathy: a 50-year odyssey. Am J Kidney Dis. 2010;56:157–67.
Lehmann S, Hoofnagle A, Hochstrasser D, Brede C, Glueckmann M, Cocho JA, et al. Quantitative clinical chemistry proteomics (qCCP) using mass spectrometry: general characteristics and application. Clin Chem Lab Med. 2013;51:919–35.
Thomas S, Hao L, Ricke WA, Li L. Biomarker discovery in mass spectrometry-based urinary proteomics. Proteomics Clin Appl. 2016;10:358–70.
Chahrour O, Cobice D, Malone J. Stable isotope labelling methods in mass spectrometry-based quantitative proteomics. J Pharm Biomed Anal. 2015;113:2–20.
Malagrino PA, Venturini G, Yogi PS, et al. Proteome analysis of acute kidney injury-Discovery of new predominantly renal candidates for biomarker of kidney disease. J Proteomics. 2017;151:66–73.
Caster DJ, Korte EA, Merchant ML, et al. Autoantibodies targeting glomerular annexin A2 identify patients with proliferative lupus nephritis. Proteomics Clin Appl. 2015;9:1012–20.
Lewandowicz A, Bakun M, Kohutnicki R, et al. Changes in urine proteome accompanying diabetic nephropathy progression. Pol Arch Med Wewn. 2015;125:27–38.
Samavat S, Kalantari S, Nafar M, et al. Diagnostic urinary proteome profile for immunoglobulin a nephropathy. Iran J Kidney Dis. 2015;9:239–48.
Integrated proteome resources. http://www.iprox.org/index. Accessed Feb 2017.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9.
Zhang M, Luo SC. Gene expression profiling of epithelial ovarian cancer reveals key genes and pathways associated with chemotherapy resistance. Genet Mol Res. 2016;15:11.
Rood IM, Merchant ML, Wilkey DW, et al. Increased expression of lysosome membrane protein 2 in glomeruli of patients with idiopathic membranous nephropathy. Proteomics. 2015;15:3722–30.
Anders HJ, Ponticelli C. Glomerular disease: membranous nephropathy and the Henle–Koch postulates. Nat Rev Nephrol. 2016;12:447–8.
Glassock RJ. Thrombo-prevention in membranous nephropathy: a new tool for decision making? Kidney Int. 2014;85:1265–6.
Huang MJ, Wei RB, Wang ZC, Xing Y, Gao YW, Li MX, et al. Mechanisms of hypercoagulability in nephrotic syndrome associated with membranous nephropathy as assessed by thromboelastography. Thromb Res. 2015;136:663–8.
de Serres F, Blanco I. Role of alpha-1 antitrypsin in human health and disease. J Intern Med. 2014;276:311–35.
Stoller JK, Aboussouan LS. A review of alpha1-antitrypsin deficiency. Am J Respir Crit Care Med. 2012;185:246–59.
Yang Y, Wei J, Huang X, Wu M, Lv Z, Tong P, et al. iTRAQ-based proteomics of chronic renal failure rats after FuShengong decoction treatment reveals haptoglobin and alpha-1-antitrypsin as potential biomarkers. Evid Based Complement Alternat Med. 2017;2017:1480514.
Maicas N, van der Vlag J, Bublitz J, Florquin S, Bakker-van Bebber M, Dinarello CA, et al. Human alpha-1-antitrypsin (hAAT) therapy reduces renal dysfunction and acute tubular necrosis in a murine model of bilateral kidney ischemia-reperfusion injury. PLoS ONE. 2017;12:e0168981.
Navarro-Munoz M, Ibernon M, Bonet J, Perez V, Pastor MC, Bayes B, et al. Uromodulin and alpha(1)-antitrypsin urinary peptide analysis to differentiate glomerular kidney diseases. Kidney Blood Press Res. 2012;35:314–25.
Choi YW, Kim YG, Song MY, et al. Potential urine proteomics biomarkers for primary nephrotic syndrome. Clin Proteomics. 2017;14:18.
Dieplinger H, Dieplinger B. Afamin-A pleiotropic glycoprotein involved in various disease states. Clin Chim Acta. 2015;446:105–10.
Lichenstein HS, Lyons DE, Wurfel MM, Johnson DA, McGinley MD, Leidli JC, et al. Afamin is a new member of the albumin, alpha-fetoprotein, and vitamin D-binding protein gene family. J Biol Chem. 1994;269:18149–54.
Zhao M, Li M, Li X, Shao C, Yin J, Gao Y. Dynamic changes of urinary proteins in a focal segmental glomerulosclerosis rat model. Proteome Sci. 2014;12:42.
Sedic M, Gethings LA, Vissers JP, Shockcor JP, McDonald S, Vasieva O, et al. Label-free mass spectrometric profiling of urinary proteins and metabolites from paediatric idiopathic nephrotic syndrome. Biochem Biophys Res Commun. 2014;452:21–6.
Kalantari S, Rutishauser D, Samavat S, Nafar M, Mahmudieh L, Rezaei-Tavirani M, et al. Urinary prognostic biomarkers and classification of IgA nephropathy by high resolution mass spectrometry coupled with liquid chromatography. PLoS ONE. 2013;8:e80830.
Authors’ contributions
LP, HXL and YL made substantial contributions to conception and design; LP and HXL were involved in the data analysis and interpretation; LP, HXL and QQL conducted the experiments and helped in drafting the manuscript or revising it critically for important intellectual content; YL and ND collected the patients samples, completed the patient database and helped in drafting the manuscript; All authors read and approved the final manuscript.
Acknowledgements
The authors gratefully acknowledge the assistance of Dr. Yan Li of Fanxing BioTech for her expertise in LC–MS/MS analysis.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files. All of the MS proteomics data have been deposited to the integrated proteome resources with the accession number IPX0001113000.
Consent for publication
Written informed consents for publication were obtained from all participants.
Ethics approval and consent to participate
This study was approved by the ethics committee of Peking University First Hospital and informed consents were obtained from all participants.
Funding
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Table S1.
Clinical characteristics of patients in group A.
Additional file 2: Table S2.
Clinical characteristics of patients in group B.
Additional file 3: Table S3.
Clinical characteristics of healthy volunteers in group C.
Additional file 4: Table S4.
Raw data of the information of 509 proteins in TMT1.
Additional file 5: Table S5.
Raw data of the information of 411 proteins in TMT2.
Additional file 6: Table S6.
The detail information of gene ontology (GO) enrichment analysis.
Additional file 7: Table S7.
The detail information of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Pang, L., Li, Q., Li, Y. et al. Urine proteomics of primary membranous nephropathy using nanoscale liquid chromatography tandem mass spectrometry analysis. Clin Proteom 15, 5 (2018). https://doi.org/10.1186/s12014-018-9183-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12014-018-9183-3