Abstract
The relationship between Neanderthals and modern humans is contentious, but recent advances in Neanderthal genomics have shed new light on their evolutionary history. Here we review the available evidence and find no indication of any Neanderthal contribution to modern genetic diversity.
Similar content being viewed by others
One of the most intriguing questions in human evolution revolves around the Neanderthals, who were the first human-like fossil species to be discovered, more than 150 years ago. What were they like and why did they disappear 30,000 years ago? Do we carry any of their genes? Three hypotheses have been proposed to explain the origin of anatomically modern humans (Homo sapiens) and their relation to so-called 'archaic' humans such as the Neanderthals (Homo neanderthalensis) (Figure 1). One is the well known 'out of Africa' or 'recent replacement' theory [1, 2]; this says that H. sapiens evolved in Africa and migrated from there relatively recently, expanding over the world and displacing those archaic humans, such as the Neanderthals, who had evolved independently in Eurasia. An older hypothesis suggests that the evolution of modern humans occurred in both Africa and Eurasia, with gene flow between the various populations; this is known as the 'multiregional' model [3–5]. A Neanderthal genome project based on DNA from fossil specimens is now under way and aims to provide us with much more information about what the Neanderthals might have been like. In particular, it should provide a definitive answer to whether there was any genetic intermixing between them and the modern humans who coexisted with them in Europe for up to 6,000 years [6] and perhaps longer in Western Asia.
Models of modern human origins. In each case, anatomically modern humans are designated in blue and Neanderthals (and other extinct Eurasian archaic human species) in red. The gray root indicates the common origin of all human species, most probably in Africa. (a) The 'African replacement' hypothesis proposes that anatomically modern humans originated in Africa, expanding into Eurasia relatively recently and replacing other human species, such as the Neanderthals, which had evolved independently there [1,2]. (b) In contrast, an older hypothesis, the 'multiregional model', envisages that the evolution of modern humans occurred in both Africa and Eurasia, maintaining local genetic continuity but with populations united by gene flow [3-6]. (c) Some researchers combine these models, seeing a recent African origin for the bulk of the human genome, but limited admixture with existing populations [48].
Phylogenetic analyses of Neanderthal DNA
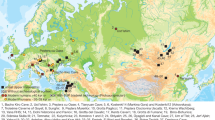
Mitochondrial DNA, which is inherited through the maternal line, has been a favored DNA sequence for determining relationships between human populations, and there is a large amount of data on the mitochondrial DNA sequences present in humans of many different ethnic groups from all over the world. In 1997, Krings and colleagues [7] first amplified and sequenced mitochondrial DNA (mtDNA) from a Neanderthal fossil – in fact, the original Neanderthal specimen. By late 2007, 14 other specimens had yielded mitochondrial sequences that could be compared (Figure 2 and Table 1). Several have yielded sequences over 300 bp long from the hypervariable region 1 of the mitochondrial control region (Figure 3). Other sequences are shorter but contain informative nucleotide positions. These mtDNA data indicate that all Neanderthal specimens sequenced up to now form a monophyletic lineage that split from the human lineage several hundred thousand years before populations of modern humans began to diverge from each other (Figure 1a).
Nevertheless, some researchers have argued that the absence of Neanderthal-related haplotypes in modern human populations does not necessarily mean that there was no interbreeding between Neanderthals and modern humans 30,000 or more years ago [8, 9]. Sequencing of mtDNA from anatomically modern human fossils 24,000 years old by Caramelli et al. [10] strongly suggested that there was no relationship with Neanderthals. But there were questions about the reliability of the DNA techniques and the possibility of contamination by DNA from those who had handled the specimens. To address such problems, Serre et al. [11] sequenced a series of Neanderthal specimens and contemporaneous early modern human fossils using Neanderthal-specific PCR primers, to avoid detecting any contaminating present-day DNA. The Neanderthal fossils yielded Neanderthal mtDNA haplotypes, but no amplifications were obtained from the well-preserved early modern samples. Serre et al. interpreted this as a significant lack of evidence of Neanderthal-modern human admixture near the time at which it may have been possible.
More recently, researchers have been successful in isolating and sequencing DNA from the Neanderthal nuclear genome. Ancient DNA entered the genomics age with the publication of around 27,000 bp of Pleistocene cave bear sequence [12] and more than 13 million bp of woolly mammoth DNA [13]. These studies used cell-based and emulsion-bead approaches to create metagenomic libraries of fossil DNA extracts [12–14]. Such libraries contain both endogenous DNA from the fossils and exogenous microbial DNA from modern contaminants and from microbes that colonized the organism after death or lived in the soil matrix. These approaches were applied to Neanderthals. A 38,000-year-old fossil from Vindija in Croatia (Vindija 80, Figure 2 and Table 1) was chosen for analysis because a preliminary PCR and subcloning of the fossil's mtDNA indicated well preserved DNA that was largely free of contamination [15]. Noonan et al. [16] obtained 65,250 bp of Neanderthal genomic sequence using a cell-based approach, while Green et al. [15] obtained more than 1 Mb of genomic sequence using an emulsion-bead based approach.
Both groups made alignments of their sequences with orthologous chimpanzee and human sequences and characterized the substitutions along each lineage. From these, an average sequence divergence time between Neanderthals and modern humans could be calculated. This parameter does not, however, necessarily measure the time that the two populations actually split. To estimate that, the two groups compared their Neanderthal sequence with information on single nucleotide polymorphisms (SNPs) in present-day humans collected by the HapMap project [17]. If the split between humans and Neanderthals is ancient, Neanderthals should rarely, or almost never, carry the 'derived' variant of a human SNP – that is, a variant that is present in some modern human lineages but not in the ancestral human lineage from which both Neanderthals and modern humans descend. On the other hand, if the split is recent, derived variants will be common in the Neanderthal genome and we should expect alleles to be shared between modern Europeans and Neanderthals.
Although they were working with DNA from the same specimen, the two teams came to very different conclusions. Noonan et al. [16] arrived at an average divergence time between Neanderthals and humans of 706,000 years and an estimated time for a population split at 370,000 years ago. They found derived human SNP variants at only three sites in the Neanderthal DNA, two of which are only found in sub-Saharan Africans and not in Europeans. They concluded that the Neanderthal contribution to modern genetic diversity was zero. Green and colleagues [15], on the other hand, calculated the average sequence divergence time between Neanderthals and humans as 516,000 years. To check whether this degree of divergence is comparable to that found within humans, they resequenced a modern human using an identical approach and compared the data to the chimp and human reference genomes. They found the average sequence divergence time between the resequenced human and the reference genome to be 459,000 years. And when Green et al. compared their Neanderthal sequence with the corresponding HapMap data, they found that around 30% of the SNPs were of the derived human type. They therefore concluded that a single ancient split between Neanderthals and humans is unlikely, and there must have been some level of recent gene flow.
Such conflicting conclusions from the same DNA sample not surprisingly led to a reanalysis of the data. Contaminating modern DNA should be less fragmented than genuine ancient DNA. To check their data for evidence of contamination, Noonan et al. [15] had compared their long sequence reads to their short sequence reads and confirmed an equal sequence divergence from modern humans across their data, indicating the absence of contamination. Green et al. [14] had not taken this step. Wall and Kim [18] reanalyzed Green et al.'s dataset and found that their long sequence reads showed significantly lower sequence divergence from modern humans than their short sequence reads, and that their short sequence reads showed an indistinguishable level of sequence divergence from Noonan et al.'s data. Wall and Kim concluded that the sequence used by Green et al. had been contaminated by human DNA – and, using a maximum likelihood analysis, estimated the contamination to be as high as 78%. We also note that Noonan et al. found that 1.3% of their metagenomic library was Neanderthal in origin, whereas Green et al. found 6.2% to be Neanderthal. If this difference is due to contamination, then it is in close agreement with Wall and Kim's likelihood estimates. We believe these findings serve as a cautionary tale that even with extremely stringent protocols, contamination of fossils with modern human DNA will remain a problem.
From sequence to function
The analysis of Neanderthal genomic DNA took an exciting step forward recently with the sequencing of two protein-coding genes that are known to have undergone adaptive evolution along the human lineage. The first gene, forkhead box 2 (FOXP2), is thought to be involved in language development, whereas the second, melanocortin 1 receptor (MC1R), is involved in skin and hair pigmentation.
Krause and colleagues [19] targeted the FOXP2 sequence in two Neanderthal specimens from El Sidron, Spain (Figure 2), excavated under sterile conditions to avoid contamination, and recovered the derived form of FOXP2 identical to that found in humans. These researchers largely ruled out human contamination through multiple control PCRs designed to detect it, and by several independent replications of the sequencing result. It has been suggested that the findings of Krause et al. mean that Neanderthals had a language ability similar to our own, though we feel that this interpretation is premature. Because no study of ancient DNA has demonstrated gene flow between Neanderthals and modern humans, Krause et al. conclude that selection fixed this variant of FOXP2 before the separation of the Neanderthal and modern human lineages.
Several genes are associated with variation in skin and hair pigmentation in humans [20–22]. MC1R affects skin color by regulating the expression of the darker eumelanin, and thus altering its ratio to the lighter pheomelanin. Low-activity variants of MC1R, for example, produce low ratios of eumelanin to pheomelanin, giving pale skin and blond to ginger hair (reviewed in [23]). Lalueza-Fox and colleagues [24] sequenced a 128 bp fragment of MC1R in two Neanderthal DNA samples; an additional one from El Sidron and one from Monti Lessini in Italy (Figure 2). They found an A to G transition, resulting in an arginine to glycine substitution, in both samples. This substitution is not found in humans and is likely to be a legitimate Neanderthal difference because A to G transitions are not typical artifacts of DNA degradation. These results were also replicated in multiple PCR experiments and in different labs.
Lalueza-Fox et al. [24] also took the unprecedented step of exploring the phenotypic effects of a Neanderthal sequence by expressing the Neanderthal MC1R, a human ancestral high-activity allele, and a derived human low-activity allele in cell culture. They found that the Neanderthal MC1R had 40% the activity of the ancestral allele, and was indistinguishable in its effects from the low-activity allele found in some modern Europeans. This study represents the first functional study of Neanderthal DNA and strongly suggests that at least some Neanderthals had pale skin and ginger hair.
The relationship between Neanderthals and modern humans
These new developments in Neanderthal genomics allow us to re-evaluate both the possible Neanderthal contribution to modern gene pools and the modern human contribution to the Neanderthal gene pool. The distribution of genetic variation within present-day humans has been interpreted to support both the recent replacement and the multiregional evolution hypotheses (Figure 1), though a consensus is developing in favor of recent replacement.
Many authors have interpreted the very recent origin of human mitochondrial DNA [25, 26], Y chromosomes [27, 28] and other loci [29, 30] in Africa as evidence for the recent replacement hypothesis. However, Templeton [31, 32] argues that there are loci that do not show a recent origin in Africa and that human history is best characterized by several population expansions and continued gene flow between modern and archaic groups. Wall [33] has argued that the data so far are insufficient to answer the question, and that sequence data from between 50 and 100 independently segregating loci in present-day humans will be required. Fagundes et al. [34] have now used data from 50 human genomic loci to compare several versions of the multiregional model with the recent replacement model by a likelihood-based approach and found that recent replacement best fits the data, with a posterior probability of 78%. The best estimate from these data is that modern humans arose around 141,000 years ago in Africa, with migration out of Africa around 51,000 years ago, which is in broad agreement with most interpretations of the fossil record [2].
The findings from Neanderthal DNA sequence fit nicely with the analysis of Fagundes et al. [34]. First, analysis of mtDNA from multiple Neanderthal samples has revealed a monophyletic origin for the Neanderthal lineage that falls outside the range of diversity of both present-day and fossil modern humans. It has been pointed out that the mitochondrial data alone were insufficient to definitively exclude the possibility of genetic admixture between modern humans and Neanderthals [11, 33, 35]. To address this question, Currat and Excoffier [36] simulated admixture between Neanderthals and modern humans during the expansion of modern humans into Europe and found that even modest amounts of mixing would result in the complete replacement of the invading modern human mtDNA by endemic Neanderthal mtDNA. Furthermore, they found that even very low levels of admixture would result in a significant minority of Neanderthal mtDNA in extant European populations. They estimate the maximum amount of admixture possible to observe no surviving Neanderthal mtDNA to be less than 0.1%, with no more than 120 admixture events during 12,000 years of overlap.
As long as mitochondria evolve neutrally, the analysis by Currat and Excoffier [36] effectively eliminates the possibility of female-mediated neutral gene flow from Neanderthals to modern humans. This leaves open the formal, although unlikely, possibility of strictly male-mediated gene flow from Neanderthals to modern humans, or the possibility of active selection against Neanderthal mtDNA. Under either of these scenarios there should be evidence for derived Neanderthal nuclear genes in modern populations. But, excluding contamination, the four studies of nuclear DNA reviewed above have all failed to show any contribution, despite the sampling of many independent loci.
It is now clear that the level of interbreeding between the two populations, if any, was so low that we are unlikely to find any neutrally evolving Neanderthal alleles in modern populations. However, it is possible that low levels of interbreeding could have led to the adaptive transfer of some alleles between species (introgression). Beneficial alleles can persist in interspecific hybrids even when the hybrids are less fit than either parent population as long as the hybrids are fertile [37]. As hybrids back-cross to a parent population, most introduced alleles will be lost to drift or to negative selection; some beneficial alleles, however, may be maintained in subsequent generations. Claims have been made for adaptive introgression from Neanderthals into populations of modern humans at the microcephalin [38] and the tau [39] loci. Some proponents of the multiregional model have gone so far as to suggest that adaptive introgression was a primary source of beneficial alleles during the evolution of modern humans [40]. While we regard this latter idea as unsupported by the available Neanderthal and human genome sequences, it is worth considering the possibility that a very limited amount of adaptive introgression has occurred.
MC1R is a good a priori candidate for adaptive introgression. It is thought that light skin is favored in Europe as a compromise between the need for vitamin D synthesis and the need to prevent folate photolysis, both caused by UV radiation [41]. Several genes affecting skin color are known to have been positively selected in European populations [21, 22], though studies of MC1R evolution have come to different conclusions [22, 42, 43]. Jolly has pointed out that the easiest way for early modern humans entering Europe to evolve light skin would be to acquire the necessary genes from Neanderthals rather than to evolve them de novo [44]. If the low-activity MC1R variant is positively selected in Europe, then MC1R presents a good opportunity to test for evidence of adaptive introgression from Neanderthals to modern humans. However, although Neanderthals and modern Europeans share a low-activity MC1R phenotype, the genotype is different (see above), which argues against significant adaptive introgression. The hypothesis could be tested more rigorously using Neanderthal sequence from other loci affecting skin color with a clearer signal of positive selection in Europeans. The failure to find evidence for adaptive introgression would strongly suggest some pre- or post-zygotic barrier to gene flow such as chromosomal incompatibility [45].
Some studies claim evidence of shared morphologies between archaic and modern humans that demonstrates gene flow between the groups ([46, 47] but see [48]). The only shared phenotype between Neanderthals and modern humans for which we know the genotype (that is, MC1R) has resulted from convergence. It should not be surprising that populations with largely similar genomes living in largely similar environments will sometimes solve evolutionary problems in largely similar ways. In light of the failure to find a Neanderthal contribution to modern gene pools, convergence ought to be considered the null hypothesis with regard to phenotypic similarity between Neanderthals and modern humans.
We interpret the findings discussed above as effectively eliminating the multiregional model of evolution with respect to Neanderthals. It seems unlikely that Neanderthals contributed any substantial fraction of modern variation and it remains to be seen whether any adaptive alleles crossed the human-Neanderthal species boundary. The continuation of the Neanderthal genome project, along with a better understanding of modern genomic diversity, will shed even more light on the origins of modern humans.
References
Howells WW: Explaining modern man: evolutionists versus migrationists. J Hum Evol. 1976, 5: 477-495. 10.1016/0047-2484(76)90088-9.
Stringer CB, Andrews P: Genetic and fossil evidence for the origin of modern humans. Science. 1988, 239: 1263-1268. 10.1126/science.3125610.
Weidenreich F: Apes, Giants and Man. 1946, Chicago: University of Chicago Press
Wolpoff MH, Zhi WX, Thorne AG: Modern Homo sapiens origins: a general theory of hominid evolution involving the fossil evidence from East Asia. The Origins of Modern Humans: A World Survey of the Fossil Evidence. Edited by: Smith FH, Spencer F. 1984, New York: Alan Liss, 411-483.
Wolpoff MH: Neandertals: not so fast. Science. 1998, 282: 1991-10.1126/science.282.5396.1991b.
Mellars P: A new radiocarbon revolution and the dispersal of modern humans in Eurasia. Nature. 2006, 439: 931-935. 10.1038/nature04521.
Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pääbo S: Neandertal DNA sequences and the origin of modern humans. Cell. 1997, 90: 19-30. 10.1016/S0092-8674(00)80310-4.
Nordborg M: On the probability of Neanderthal ancestry. Am J Hum Genet. 1998, 63: 1237-1240. 10.1086/302052.
Relethford JH: Absence of regional affinities of Neandertal DNA with living humans does not reject multiregional evolution. Am J Phys Anthropol. 2001, 115: 95-98. 10.1002/ajpa.1060.
Caramelli D, Lalueza-Fox C, Vernesi C, Lari M, Casoli A, Mallegni F, Chiarelli B, Dupanloup I, Bertranpetit J, Barbujani G, Bertorelle G: Evidence for a genetic discontinuity between Neanderthals and 24,000-year-old anatomically modern Europeans. Proc Natl Acad Sci USA. 2003, 100: 6593-6597. 10.1073/pnas.1130343100.
Serre D, Langaney A, Chech M, Teschler-Nicola M, Paunovic M, Mennecier P, Hofreiter M, Possnert G, Pääbo S: No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol. 2004, 2: E57-10.1371/journal.pbio.0020057.
Noonan JP, Hofreiter M, Smith D, Priest JR, Rohland N, Rabeder G, Krause J, Detter JC, Pääbo S, Rubin EM: Genomic sequencing of Pleistocene cave bears. Science. 2005, 309: 597-599. 10.1126/science.1113485.
Poinar HN, Schwarz C, Qi J, Shapiro B, Macphee RD, Buigues B, Tikhonov A, Huson DH, Tomsho LP, Auch A, Rampp M, Miller W, Schuster SC: Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006, 311: 392-394. 10.1126/science.1123360.
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, et al: Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005, 437: 376-380.
Green RE, Krause J, Ptak SE, Briggs AW, Ronan MT, Simons JF, Du L, Egholm M, Rothberg JM, Paunovic M, Pääbo S: Analysis of one million base pairs of Neanderthal DNA. Nature. 2006, 444: 330-336. 10.1038/nature05336.
Noonan JP, Coop G, Kudaravalli S, Smith D, Krause J, Alessi J, Chen F, Platt D, Pääbo S, Pritchard JK, Rubin EM: Sequencing and analysis of Neanderthal genomic DNA. Science. 2006, 314: 1113-1118. 10.1126/science.1131412.
HapMap. [http://www.hapmap.org]
Wall JD, Kim SK: Inconsistencies in Neanderthal genomic DNA sequences. PLoS Genet. 2007, 3: e175-10.1371/journal.pgen.0030175.
Krause J, Lalueza-Fox C, Orlando L, Enard W, Green RE, Burbano HA, Hublin JJ, Hänni C, Fortea J, de la Rasilla M, Bertranpetit J, Rosas A, Pääbo S: The derived FOXP2 variant of modern humans was shared with Neandertals. Curr Biol. 2007, 17: 1908-1912. 10.1016/j.cub.2007.10.008.
McEvoy B, Beleza S, Shriver MD: The genetic architecture of normal variation in human pigmentation: an evolutionary perspective and model. Hum Mol Genet. 2006, 15 (Spec No 2): R176-R181. 10.1093/hmg/ddl217.
Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, Manolescu A, Karason A, Palsson A, Thorleifsson G, Jakobsdottir M, Steinberg S, Pálsson S, Jonasson F, Sigurgeirsson B, Thorisdottir K, Ragnarsson R, Benediktsdottir KR, Aben KK, Kiemeney LA, Olafsson JH, Gulcher J, Kong A, Thorsteinsdottir U, Stefansson K: Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007, 39: 1443-1452. 10.1038/ng.2007.13.
Norton HL, Kittles RA, Parra E, McKeigue P, Mao X, Cheng K, Canfield VA, Bradley DG, McEvoy B, Shriver MD: Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol. 2007, 24: 710-722. 10.1093/molbev/msl203.
Rees JL: Genetics of hair and skin color. Annu Rev Genet. 2003, 37: 67-90. 10.1146/annurev.genet.37.110801.143233.
Lalueza-Fox C, Römpler H, Caramelli D, Stäubert C, Catalano G, Hughes D, Rohland N, Pilli E, Longo L, Condemi S, de la Rasilla M, Fortea J, Rosas A, Stoneking M, Schöneberg T, Bertranpetit J, Hofreiter M: A melanocortin 1 receptor allele suggests varying pigmentation among Neanderthals. Science. 2007, 318: 453-455. 10.1126/science.1147417.
Cann RL, Stoneking M, Wilson AC: Mitochondrial DNA and human evolution. Nature. 1987, 325: 31-36. 10.1038/325031a0.
Ingman M, Kaessmann H, Pääbo S, Gyllensten U: Mitochondrial genome variation and the origin of modern humans. Nature. 2000, 408: 708-713. 10.1038/35047064.
Hammer MF, Karafet T, Rasanayagam A, Wood ET, Altheide TK, Jenkins T, Griffiths RC, Templeton AR, Zegura SL: Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol Biol Evol. 1998, 15: 427-441.
Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonné-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ: Y chromosome sequence variation and the history of human populations. Nat Genet. 2000, 26: 358-361. 10.1038/81685.
Tishkoff SA, Dietzsch E, Speed W, Pakstis AJ, Kidd JR, Cheung K, Bonne-Tamir B, Santachiara-Benerecetti AS, Moral P, Krings M: Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996, 271: 1380-1387. 10.1126/science.271.5254.1380.
Tishkoff SA, Pakstis AJ, Stoneking M, Kidd JR, Destro-Bisol G, Sanjantila A, Lu RB, Deinard AS, Sirugo G, Jenkins T, Kidd KK, Clark AG: Short tandem-repeat polymorphism/alu haplotype variation at the PLAT locus: implications for modern human origins. Am J Hum Genet. 2000, 67: 901-925. 10.1086/303068.
Templeton AR: Haplotype trees and modern human origins. Am J Phys Anthropol. 2005, 33-59. 10.1002/ajpa.20351. Suppl 41
Templeton AR: Genetics and recent human evolution. Evolution: Int J Org Evol. 2007, 61: 1507-1519.
Wall JD: Detecting ancient admixture in humans using sequence polymorphism data. Genetics. 2000, 154: 1271-1279.
Fagundes NJ, Ray N, Beaumont M, Neuenschwander S, Salzano FM, Bonatto SL, Excoffier L: Statistical evaluation of alternative models of human evolution. Proc Natl Acad Sci USA. 2007, 104: 17614-17619. 10.1073/pnas.0708280104.
Wall JD, Hammer MF: Archaic admixture in the human genome. Curr Opin Genet Dev. 2006, 16: 606-610. 10.1016/j.gde.2006.09.006.
Currat M, Excoffier L: Modern humans did not admix with Neanderthals during their range expansion into Europe. PLoS Biol. 2004, 2: e421-10.1371/journal.pbio.0020421.
Arnold ML, Bulger MR, Burke JM, Hempel AL, Williams JH: Natural hybridization: how long can you go and still be important?. Ecology. 1999, 80: 371-381.
Evans PD, Mekel-Bobrov N, Vallender EJ, Hudson RR, Lahn BT: Evidence that the adaptive allele of the brain size gene microcephalin introgressed into Homo sapiens from an archaic Homo lineage. Proc Natl Acad Sci USA. 2006, 103: 18178-18183. 10.1073/pnas.0606966103.
Hardy J, Pittman A, Myers A, Gwinn-Hardy K, Fung HC, de Silva R, Hutton M, Duckworth J: Evidence suggesting that Homo neanderthalensis contributed the H2 MAPT haplotype to Homo sapiens. Biochem Soc Trans. 2005, 33: 582-585. 10.1042/BST0330582.
Hawks J, Cochran G: Dynamics of adaptive introgression from archaic to modern humans. PaleoAnthropology. 2006, 2006: 101-115.
Jablonski NG, Chaplin G: The evolution of human skin coloration. J Hum Evol. 2000, 39: 57-106. 10.1006/jhev.2000.0403.
Rana BK, Hewett-Emmett D, Jin L, Chang BH, Sambuughin N, Lin M, Watkins S, Bamshad M, Jorde LB, Ramsay M, Jenkins T, Li WH: High polymorphism at the human melanocortin 1 receptor locus. Genetics. 1999, 151: 1547-1557.
Harding RM, Healy E, Ray AJ, Ellis NS, Flanagan N, Todd C, Dixon C, Sajantila A, Jackson IJ, Birch-Machin MA, Rees JL: Evidence for variable selective pressures at MC1R. Am J Hum Genet. 2000, 66: 1351-1361. 10.1086/302863.
Jolly CJ: A proper study for mankind: analogies from the Papionin monkeys and their implications for human evolution. Am J Phys Anthropol. 2001, 116: 177-204. 10.1002/ajpa.10021.
Disotell TR: 'Chumanzee' evolution: the urge to diverge and merge. Genome Biol. 2006, 7: 240-10.1186/gb-2006-7-11-240.
Wolpoff MH, Hawks J, Frayer DW, Hunley K: Modern human ancestry at the peripheries: a test of the replacement theory. Science. 2001, 291: 293-297. 10.1126/science.291.5502.293.
Hawks J, Oh S, Hunley K, Dobson S, Cabana G, Dayalu P, Wolpoff MH: An Australasian test of the recent African origin theory using the WLH-50 calvarium. J Hum Evol. 2000, 39: 1-22. 10.1006/jhev.1999.0384.
Brauer G, Collard M, Stringer C: On the reliability of recent tests of the Out of Africa hypothesis for modern human origins. Anat Rec. 2004, 279: 701-707. 10.1002/ar.a.20064.
Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pääbo S: Neandertal DNA sequences and the origin of modern humans. Cell. 1997, 90: 19-30. 10.1016/S0092-8674(00)80310-4.
Krings M, Geisert H, Schmitz RW, Krainitzki H, Pääbo S: DNA sequence of the mitochondrial hypervariable region II from the Neandertal type specimen. Proc Natl Acad Sci USA. 1999, 96: 5581-5585. 10.1073/pnas.96.10.5581.
Schmitz RW, Serre D, Bonani G, Feine S, Hillgruber F, Krainitzki H, Pääbo S, Smith FH: The Neandertal type site revisited: interdisciplinary investigations of skeletal remains from the Neander Valley, Germany. Proc Natl Acad Sci USA. 2002, 99: 13342-13347. 10.1073/pnas.192464099.
Ovchinnikov IV, Gotherstrom A, Romanova GP, Kharitonov VM, Liden K, Goodwin W: Molecular analysis of Neanderthal DNA from the northern Caucasus. Nature. 2000, 404: 490-493. 10.1038/35006625.
Krings M, Capelli C, Tschentscher F, Geisert H, Meyer S, von Haeseler A, Grossschmidt K, Possnert G, Paunovic M, Pääbo S: A view of Neandertal genetic diversity. Nat Genet. 2000, 26: 144-146. 10.1038/79855.
Beauval C, Maureille B, Lacrampe-Cuyaubère F, Serre D, Peressinotto D, Bordes JG, Cochard D, Couchoud I, Dubrasquet D, Laroulandie V, Lenoble A, Mallye JB, Pasty S, Primault J, Rohland N, Pääbo S, Trinkaus E: A late Neandertal femur from Les Rochers-de-Villeneuve, France. Proc Natl Acad Sci USA. 2005, 102: 7085-7090. 10.1073/pnas.0502656102.
Orlando L, Darlu P, Toussaint M, Bonjean D, Otte M, Hanni C: Revisiting Neandertal diversity with a 100,000 year old mtDNA sequence. Curr Biol. 2006, 16: R400-R402. 10.1016/j.cub.2006.05.019.
Caramelli D, Lalueza-Fox C, Capelli C, Lari M, Sampietro ML, Gigli E, Milani L, Pilli E, Guimaraes S, Chiarelli B, Marin VT, Casoli A, Stanyon R, Bertranpetit J, Barbujani G: A highly divergent mtDNA sequence in a Neandertal individual from Italy. Curr Biol. 2006, 16: R630-R632. 10.1016/j.cub.2006.07.043.
Lalueza-Fox C, Sampietro ML, Caramelli D, Puder Y, Lari M, Calafell F, Martínez-Maza C, Bastir M, Fortea J, de la Rasilla M, Bertranpetit J, Rosas A: Neandertal evolutionary genetics: mitochondrial DNA data from the iberian peninsula. Mol Biol Evol. 2005, 22: 1077-1081. 10.1093/molbev/msi094.
Lalueza-Fox C, Krause J, Caramelli D, Catalano G, Milani L, Sampietro ML, Calafell F, Martínez-Maza C, Bastir M, García-Tabernero A, de la Rasilla M, Fortea J, Pääbo S, Bertranpetit J, Rosas A: Mitochondrial DNA of an Iberian Neandertal suggests a population affinity with other European Neanderthals. Curr Biol. 2006, 16: R629-R630. 10.1016/j.cub.2006.07.044.
Krause J, Orlando L, Serre D, Viola B, Prufer K, Richards MP, Hublin JJ, Hanni C, Derevianko AP, Pääbo S: Neanderthals in central Asia and Siberia. Nature. 2007, 449: 902-904. 10.1038/nature06193.
Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG: Sequence and organization of the human mitochondrial genome. Nature. 1981, 290: 457-465. 10.1038/290457a0.
Acknowledgements
We would like to thank Cliff Jolly, Randy White, Andy Burrell, Tony Tosi and Robin Allaby for useful comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
About this article
Cite this article
Hodgson, J.A., Disotell, T.R. No evidence of a Neanderthal contribution to modern human diversity. Genome Biol 9, 206 (2008). https://doi.org/10.1186/gb-2008-9-2-206
Published:
DOI: https://doi.org/10.1186/gb-2008-9-2-206