Abstract
Background
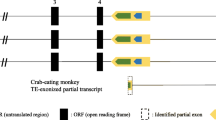
SP100 is a nuclear protein that displays a number of alternative splice variants. In Old World monkeys, apes and humans one of these variants is extended by a retroprocessed pseudogene, HMG1L3, whose antecedent gene is a member of the family of high-mobility-group proteins, HMG1. This is one of only a few documented cases of a retropseudogene being incorporated into another gene as a functional exon. In addition to the HMG1L3 insertion, Old World monkey genomes also contain an Alu sequence within the last SP100-HMG intron. PCR amplification of the 3' end of the SP100 gene using genomic DNAs from human and New World and Old World monkey species, followed by direct sequencing of the amplicons has made dating the HMG1L3 and Alu insertion events possible.
Results
PCR amplifications confirm that the HMG1L3 retrotransposition into the SP100 locus occurred after divergence of New World and Old World monkey lineages, some 35-40 million years ago. PCR amplification also shows that an upstream Alu sequence was inserted in the last SP100-HMG intron after divergence of the Old World monkey and ape lineages. Direct sequencing of the Alu in five Old World monkey species places the latter event at around 19 million years ago. Finally, ten single base mutations and one deletion in the Alu differentiate African from Asian Old World monkey species.
Conclusions
PCR and DNA sequence analysis of 'genetic fossils' such as retropseudogenes and Alu elements in primates give details as to the timing of such events and can reveal sequence features useful for other molecular phylogenetic applications.




Similar content being viewed by others
References
Vanin EF: Processed pseudogenes: characteristics and evolution. Annu Rev Genet. 1985, 19: 253-272. 10.1146/annurev.ge.19.120185.001345.
Mighell AJ, Smith NR, Robinson PA, Markham AF: Vertebrate pseudogenes. FEBS Lett. 2000, 468: 109-114. 10.1016/S0014-5793(00)01199-6.
Goncalves I, Duret L, Mouchiroud D: Nature and structure of human genes that generate retropseudogenes. Genome Res. 2000, 10: 672-678. 10.1101/gr.10.5.672.
Gojobori T, Li W-H, Graur D: Patterns of nucleotide substitution in pseudogenes and functional genes. J Mol Evol. 1982, 18: 360-369.
Ophir R, Graur D: Patterns and rates of indel evolution in processed pseudogenes from humans and murids. Gene. 1997, 205: 191-202. 10.1016/S0378-1119(97)00398-3.
Casane D, Boissinot S, Chang BH, Shimmin LC, Li W: Mutation pattern variation among regions of the primate genome. J Mol Evol. 1997, 45: 216-226.
Devor EJ: Use of molecular beacons to verify that the serine hydroxy-methyltransferase pseudogene SHMT-ps1 is unique to the order Primates. Genome Biol. 2001, 2: research0006.1-0006.5. 10.1186/gb-2001-2-2-research0006.
Devor EJ, Dill-Devor RM, Magee HJ, Waziri R: Serine hydroxymethyltransferase pseudogene SHMT-ps1: A unique genetic marker of the order Primates. J Exp Zool. 1998, 282: 150-156. 10.1002/(SICI)1097-010X(199809/10)282:1/2<150::AID-JEZ16>3.3.CO;2-L.
Pompei F, Ciminelli BM, Modiano G: Two ethnic-specific polymorphisms in the human beta pseudogene of hemoglobin. Hum Biol. 1998, 70: 659-666.
Boyson JE, Iwanaga KK, Urvater JA, Hughes AL, Golos TG, Watkins DI: Evolution of a new nonclassical MHC class I locus in two Old World primate species. Immunogenetics. 1999, 49: 86-98. 10.1007/s002510050467.
Brosius J: Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica. 1999, 107: 209-238. 10.1023/A:1004018519722.
Seeler JS, Marchio A, Sitterlin D, Transy C, Dejean A: Interaction of SP100 with HP1 proteins: A link between the promyeloctyic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci USA. 1998, 95: 7316-7321. 10.1073/pnas.95.13.7316.
Rogalla P, Borda Z, Meyer-Bolte K, Tran KH, Hauke S, Nimzyk R, Bullerdiek J: Mapping and molecular characterization of five HMG1-related DNA sequences. Cytogenet Cell Genet. 1998, 83: 124-129. 10.1159/000015147.
Rogalla P, Kazmierczak B, Flohr AM, Hauke S, Bullerdiek J: Back to the roots of a new exon - the molecular archaeology of a SP100 splice variant. Genomics. 2000, 63: 117-122. 10.1006/geno.1999.6008.
Kay RF, Ross C, Williams BA: Anthropoid origins. Science. 1997, 275: 797-804. 10.1126/science.275.5301.797.
Hamdi H, Nishio H, Zielinski R, Dugaiczyk A: Origin and phylogenetic distribution of Alu DNA repeats: Irreversible events in the evolution of primates. J Mol Biol. 1999, 289: 861-871. 10.1006/jmbi.1999.2797.
Minghetti PP, Dugaiczyk A: The emergence of new DNA repeats and the divergence of primates. Proc Natl Acad Sci USA. 1993, 90: 1872-1876.
Miura O, Sagahara Y, Nakamura Y, Hirosawa S, Aoki N: Restriction fragment length polymorphism caused by a deletion involving Alu sequences within the human α2-plasmin inhibitor gene. Biochemistry. 1989, 28: 4934-4938.
Edwards MC, Gibbs RA: A human dimorphism resulting from loss of an Alu. Genomics. 1992, 14: 590-597.
Britten RJ, Baron WF, Stout DB, Davidson EH: Sources and evolution of human Alu repeated sequences. Proc Natl Acad Sci USA. 1988, 85: 4770-4774.
Batzer MA, Deininger PL, Hellmann-Blumberg U, Jurka J, Labauda D, Rubin CM, Schmid CW, Zietkiewicz E, Zuckerkandl E: Standardized nomenclature for Alu repeats. J Mol Evol. 1996, 42: 3-6.
Disotell TR, Honeycutt RL, Ruvolo M: Mitochondrial DNA phylogeny of the Old-World monkey tribe Papionini. Mol Biol Evol. 1992, 9: 1-13.
Morales JC, Melnick DJ: Phylogenetic relationships of the macaques (Cercopithecidae: Macaca), as revealed by high resolution restrcition site mapping of mitochondrial ribsomal genes. J Hum Evol. 1998, 34: 1-23. 10.1006/jhev.1997.0171.
Szalay FS, Delson E: Evolutionary History of the Primates. New York: Academic Press;. 1979
Miyata T, Yasunaga T: Rapidly evolving mouse alpha-globin-related pseudogene and its evolutionary history. Proc Natl Acad Sci USA. 1981, 78: 450-453.
Acknowledgements
I thank Moses Schanfield, Edward Max, Boris Lapin and the Southwest Foundation for Biomedical Research for their generosity in providing genomic DNA samples. Amplicon sequencing was carried out by Susanna Rezikyan at IDT.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Devor, E.J. Molecular archeology of an SP100 splice variant revisited: dating the retrotranscription and Aluinsertion events. Genome Biol 2, research0040.1 (2001). https://doi.org/10.1186/gb-2001-2-9-research0040
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/gb-2001-2-9-research0040