Abstract
Tumor necrosis factor α (TNF-α)is a host inflammatory factor. Bacteria increase TNF-α expression in a variety of human diseases including infectious diseases, inflammatory bowel diseases, and cancer. It is unknown, however, how TNF-α directly modulates bacterial protein expression during intestinal infection and chronic inflammation. In the current study, we hypothesize that Salmonella typhimurium senses TNF-α and show that TNF-α treatment modulates Salmonella virulent proteins (called effectors), thus changing the host-bacterial interaction in intestinal epithelial cells. We investigated the expression of 23 Salmonella effectors after TNF-α exposure. We found that TNF-α treatment led to differential effector expression: effector SipA was increased by TNF-α treatment, whereas the expression levels of other effectors, including gogB and spvB, decreased in the presence of TNF-α. We verified the protein expression of Salmonella effectors AvrA and SipA by Western blots. Furthermore, we used intestinal epithelial cells as our experimental model to explore the response of human intestinal cells to TNF-α pretreated Salmonella. More bacterial invasion was found in host cells colonized with Salmonella strains pretreated with TNF-α compared to Salmonella without TNF-α treatment. TNF-α pretreated Salmonella induced higher proinflammatory JNK signalling responses compared to the Salmonella strains without TNF-α exposure. Exposure to TNF-α made Salmonella to induce more inflammatory cytokine IL-8 in intestinal epithelial cells. JNK inhibitor treatment was able to suppress the effects of TNF-pretreated-Salmonella in enhancing expressions of phosphorylated-JNK and c-jun and secretion of IL-8. Overall, our study provides new insights into Salmonella-host interactions in intestinal inflammation.
Similar content being viewed by others
Background
Tumor necrosis factor α(TNF-α)is a pleiotropic inflammatory cytokine with increased expression in many human diseases. These diseases include septic shock, cancer, AIDS, multiple sclerosis, diabetes, rheumatoid arthritis, and inflammatory bowel disease [1–6]. It is well documented that multiple factors from bacteria, viruses, and parasites stimulate production of TNF-α in the host [7–10]. Hence, in hosts with inflammatory diseases, enteric bacteria are potentially exposed to high levels of TNF-α.
Bacteria can sense signal molecules secreted by their hosts. This communication mechanism between bacterium is called "quorum sensing" (QS) [11, 12]. QS utilizes hormone-like compounds referred to as autoinducers to regulate bacterial gene expression [13, 14]. QS also applies to the communication between the host and bacteria [11]. However, it is unknown how TNF-α from host cells directly modulates bacterial protein expression during infection and chronic inflammation.
Salmonella is a leading cause of gastrointestinal disease worldwide. Salmonella uses the type three secretion system (TTSS), a needle-like protein transport device to inject virulence proteins into eukaryotic host cells. These virulence factors, called effectors, paralyze or reprogram the eukaryotic cell to the benefit of the pathogen [15–17]. The activity of TTSS effectors allows bacteria to invade non-phagocytic cells or inhibit phagocytosis, regulate pro-inflammatory responses, prevent autophagy, or modulate intracellular trafficking [18]. Salmonella effectors display a large repertoire of biochemical activities and modulate the function of crucial host regulatory molecules[19–22].
Effectors are encoded via specific pathogenicity island 1 (SPI-1) and 2 (SPI-2). Over 30 Salmonella effectors, including AvrA, SipA, SipB, Gog B, and SpVB, have been shown to manipulate a succession of key signaling transduction pathways and physiological functions of host cells [19]. AvrA, SipA, SipB, SopB, SopD, SopE, SopE2 are SPI-1 effectors. SipA, SipB, SopB, SopD, SopE, SopE2 and other effectors are known to induce membrane deformation and ruffling that triggers bacterial internalization, promoting invasion [19, 23, 24]. The SPI-2 effectors, such as Gog B and SpVB, promote bacterial replication and systemic spread [19–22]. Recent studies indicate that there may be interplay between SPI-1 and SPI-2 effectors [19]. Although Salmonella is one of the best characterized pathogens, it remains unknown how virulence effector gene expression changes in response to host factors, such as TNF-α.
In Salmonella strains, AvrA is an acid-inducible effector that is strongly correlated with food hygiene and food-borne infection [25–27]. Our publications and others' have demonstrated that AvrA is a multifunctional protein that plays a critical role in inhibiting inflammation, regulating epithelial apoptosis, and enhancing proliferation during bacterial infection [28–32]. Stimulation of inflammation by effectors is crucial for Salmonella to grow in the intestine [33]. Effectors, such as SipA, SopE, and SopB, are known to activate inflammation in host cells [24, 34–41]. Un-controlled inflammation is harmful to the host, however, and eventually damages the niche occupied by Salmonella during infection. Salmonella secreted factor L (SseL) [42–44], SspH 1 [45], SptP, and AvrA may reverse the activation of signaling pathways induced by other Salmonella effectors [19, 46, 47].
Intestinal epithelial cells are physically linked by intercellular junctional complexes that regulate multiple functions including polarity, mechanical integrity, and signaling capacity [48]. Salmonella can invade and replicate within intestinal epithelial cells during the infection process [49]. Nontyphoidal Salmonella serotypes such as Salmonella typhimurium provoke an intense intestinal inflammatory response, consisting largely of neutrophil migration across the epithelial lining of the intestine [50, 51]. Studies of S. typhimurium-infected laboratory animals and cultured epithelial cells have shown that bacteria rapidly enter epithelial cells after transient degeneration of the host cell surface microvilli and induce inflammatory responses [52–58]. Not surprisingly, the ability of S. typhimurium to enter epithelial cells constitutes a crucial step in pathogenesis. Salmonella invasion of the intestinal epithelium requires the virulence-associated TTSS [19, 28, 34, 53, 59]. Within the host intestine specialized antigen-sampling M cells, which reside in the epithelium overlying lymphoid tissues in the gut, are a preferred site of Salmonella invasion [60]. The factors involved in Salmonella-M cell interactions, however, are not well understood. Clearly, studying effectors can uncover important mechanisms of regulation in host-bacteria interaction.
A recent study demonstrated that Salmonella gastroenteritis increases short- and long-term risk of inflammatory bowel disease [61]. Chronic intestinal inflammation enhances TNF-α levels in the host [62]. Therefore, enteric Salmonella is potentially exposed to TNF-α. In the current study, we hypothesize that Salmonella senses the host inflammatory factor TNF-α and that TNF-α treatment modulates Salmonella TTSS effectors, thus changing the host-bacteria interaction. We investigated the gene expression of Salmonella effectors changed by TNF-α and responses of the human intestinal cells to TNF-α treated Salmonella. We verified the expression levels of some effector proteins by Western blots. Furthermore, we used human intestinal epithelial cells as our experimental model to explore bacterial invasion and the proinflammatory NF-κB and c-Jun N-terminal kinase (JNK) signaling pathways in response to Salmonella strains with or without TNF-α pre-treatment. We found that TNF-α treatment modulated effector expression in a differentiated manner. Salmonella strains pre-treated with TNF-α induced more bacteria internalization and a more severe inflammatory response in intestinal epithelial cells than untreated Salmonella strains. Our study provides new insights into host factor regulation of bacterial effector expression through inflammatory responses.
Materials and methods
Bacterial strains and growth conditions
Salmonella strains (listed in Table 1) include wild-type (WT), S. typhimurium ATCC 14028s, S. typhimurium PhoPC [63], Salmonella typhimurium 1344 (SL1344), and an AvrA mutant strain lacking the AvrA gene (SL1344AvrA-) (provided by Dr. Jorge Galan from Yale University) [25]. Wild-type S. typhimurium 14028s AvrA - was generated in our laboratory based on previously published methods by Hamilton et al., and Miller et al. [64, 65]. Briefly, the AvrA gene, flanked by upstream and downstream Salmonella chromosome sequences, was cloned into pMAK705 (chloramphenicol resistant). The construct plasmid was transformed into the Salmonella WT14028s strain by electroporation with a Gene Pulser apparatus (Bio-Rad, Munich, Germany) and grown at 30°C on chloramphenicol plates. Resulting colonies were then grown at 42°C to select for integrants. The integrants were subsequently grown at 30°C, the temperature at which the plasmid can leave the chromosome and autonomously replicate. AvrA gene deletion was screened by PCR. AvrA deletion was also verified by Western blot using the anti-AvrA antibody. The resulting strain was named SL14028s AvrA-.
Bacteria were grown under the following conditions: non-agitated microaerophilicbacterial cultures were prepared by inoculation of 10 ml of Luria-Bertani broth with0.01 ml of a stationary phase culture with or without TNF-α (10 ng/ml), followed by overnight incubation(~18 h) at 37°C, as previously described [53]. Overnight cultures of bacteria were concentrated 33-fold in Hank's balanced salt solution (HBSS) supplemented with 10 mM HEPES, pH 7.4. The overnight cultures from the TNF-α pretreated Salmonella strains were washed thoroughly with HBSS 3 times to get rid of potential TNF-α residue in the media. The bacteria were then resuspended in fresh HBSS for cell lysis or colonization in the intestinal epithelial cells.
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted from bacteria using a Qiagen RNeasy mini kit (Cat: 74104. Qiagen, Valencia, CA) according to the manufacturer's protocol. Total RNA was further digested with DNase I (Cat: 18068-015. Invitrogen, Carlsbad, CA, USA). RNA integrity was verified by gel electrophoresis. Extracted RNA yield and purity was then determined by measuring absorbance in the 220 nm to 350 nm range. From the resulting spectra, the concentration of nucleic acids was estimated using the absorbance values at 260 nm, while the purity of each sample was determined by calculating the 260/280 and 260/230 ratios. RNA reverse transcription was performedusing a SuperScript III kit (Invitrogen, Cat: 18080-051)according to the manufacturer's directions. cDNA reactionproducts were then used in a quantitative PCR reaction. The reaction mixture was subjected to 29 cycles of PCR amplification using Taq polymerase (Fermentas, Glen Burnie, Maryland. Cat: EP0404). All PCR primers (Table 2) were designedusing Lasergene software (DNAStar, Madison, WI). PCR products were separated on 2% agarose gels and densitometry readings of the DNA bands were taken using a Kodak IS2000R. The densitometry value of each PCR band was detected using KODAK MI 4.0.3. All expressionlevels were normalized to the bacterial reference gene, Mdh, of the same sample, using forward (5'-ATGAAAGTCGCAGTCCTCGGCGCTGCTGGCGG-3') and reverse (5'-ATATCTTTYTTCAGCGTATCCAGCAT-3')primers for malate dehydrogenase (Mdh) [66]. All PCR reactionswere performed in triplicate. The digital images are representative of the original data.
Immunoblotting for bacterial SipA and AvrA
Bacteria were lysed in lysis buffer (50 mM Tris, pH 6.8, 100 mM dithiothreitol, 2% SDS, 0.1%bromophenol blue, 10% glycerol) and sonicated. Equal amounts of total proteins were loaded, separated by SDS-PAGE, and processed for immunoblotting with an anti-SipA antibody (generated by Dr. Ho-Young Kang, Pusan National University, Korea) or anti-AvrA antibody. For the anti-AvrA antibody, a 15-amino-acid peptide CGEEPFLPSDKADRY was designed based on AvrA amino acids 216-230. Two rabbits were injected with the peptide and a polyclonal antibody for AvrA was tested and purified, as previously described [30]. Immunoblotting was visualized by enhanced chemi-luminescence (ECL). Chemi-luminescent signals were collected and scanned from ECL Hyperfilm (Amersham Pharmacia Biotech) with a Scanjet 7400c backlit flatbed scanner (Hewlett-Packard Co., Palo Alto, CA). Bands were quantified using Kodak MI software (v.4.0.3). The digital images are representative of the original data.
Intestinal epithelial cell culture
Human colonic epithelial HCT116 cells (American Type Culture Collection, Manassas, VA) were grown in DMEM (high glucose, 4.5 g/L) supplemented with 10% (vol/vol) fetal bovine serum, 50 μg/ml streptomycin, and 50 U/ml penicillin.
S. typhimurium invasion of human epithelial monolayers
Infection of HCT116 cells was performed by a previously described method [53]. Bacterial solution (~20 bacteria/epithelial cell) was added and bacterial invasion was assessed after 1 hour. Cell-associated bacteria, representing bacteria adhered to and/or internalized into the monolayers, were released by incubation with 100 μl of 1% Triton X-100 (Sigma). Internalized bacteria were those obtained from lysis of the epithelial cells with 1% Triton X-100, 20 min after the addition of gentamicin (50 μg/ml). Gentamicin, an aminoglycoside antibiotic, does not permeate eukaryotic plasma membranes and is therefore cytolytic only to extracellular populations of bacteria while intracellular bacteria populations remain viable [67]. For both cell associated and internalized bacteria, 0.9 ml LB broth was then added and each sample was vigorously mixed and quantitated by plating for CFU on MacConkey agar medium.
Immunoblotting for epithelial cell signaling
Intestinal epithelial cells were incubated with equal numbers of the indicated S. typhimurium strain (about 20 bacteria per epithelial cell) for 30 minutes, washed, and incubated in fresh DMEM for 30 minutes as previously described [53, 68, 69]. Cells were rinsed twice in ice-cold HBSS, lysed in protein lysis buffer (50 mM Tris, pH 6.8, 100 mM dithiothreitol, 2% SDS, 0.1%bromophenol blue, 10% glycerol), and sonicated. Equal amounts of protein were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and immunoblotted with one of the following primary antibodies: anti-p65 (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), anti-IκBα, anti-JNK, anti-phospho-IκBα, anti-phospho-c-JUN (Cell Signal, Beverly, MA), or anti-β-actin (Sigma-Aldrich, Milwaukee, WI, USA) antibodies and visualized by ECL.
Real-time quantitative PCR analysis of the IL-8 mRNA
Total RNA was extracted from epithelial cell monolayers usingTRIzol reagent (Invitrogen, Carlsbad, CA). RNA integrity was verified by gel electrophoresis. RNA reverse transcription was doneusing the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA)according to the manufacturer's directions. The RT cDNA reactionproducts were subjected to quantitative real-time PCR usingthe MyiQ single-color real-time PCR detection system (Bio-Rad)and iQ SYBR green supermix (Bio-Rad) according to the manufacturer'sdirections. IL-8 cDNA was amplified by using primers to thehuman IL-8 gene that are complementary to regions in exon 1(5'-TGCATAAAGACATACTCCAAACCT) and overlapping the splice sitebetween exons 3 and 4 (5'-AATTCTCAGCCCTCTTCAAAAA). All expressionlevels were normalized to the GAPDH levels of the same sample, using forward (5-CTTCACCACCATGGAGAAGGC) and reverse (5'-GGCATGGACTGTGGTCATGAG)primers for GAPDH. Percent expression was calculated as theratio of the normalized value of each sample to that of thecorresponding untreated control cells. All real-time PCR reactionswere performed in triplicate. All PCR primers were designedusing Lasergene software (DNAStar, Madison, WI).
Salmonella-induced human IL-8 secretion
HCT116 cells were cultured in DMEM, followed by incubation in Salmonella-containing HBSS (1.6 × 1010 bacteria/ml) for 30 min, washed 3 times in HBSS, and incubated at 37°C for 6 hours. Cell supernatants were removed and assayed for IL-8 by ELISA in 96-well plates as described previously [53].
Treatment with JNK inhibitor SP600125
To determine whether the effects of TNF is required for JNK, cells were treated with a JNK inhibitor SP600125 (EMD Biosciences, San Diego, CA). SP600125 (50 μM) was added directly to the culture medium one hours before Salmonella treatment. For Western blot assay, HCT116 (with SP600125 pretreatment) were incubated with Salmonella (SP600125 50 μM) 1 hour, washed three times in HBSS and incubated HBSS (SP600125 50 μM) for 1 hour, then harvested. Levels of indicated proteins were determined by Western blotting as described above. For Salmonella invasion and IL-8 ELISA: HCT116 (with SP600125 pretreatment) were incubated with Salmonella (SP600125 50 μM) 1 hour, washed three times in HBSS and incubated DMEM for 6 hours.
Statistical analysis
Data are expressed as means ± SD. All statistical tests were 2-sided. P values of less than .05 were considered to be statistically significant. Differences between two samples were analyzed by a Student's t-test. Statistical analyses were performed using SAS version 9.2 (SAS Institute, Inc., Cary, NC).
Results
The alteration of Salmonella effector gene expression after TNF-α treatment
We first tested whether TNF-α treatment changes the mRNA expression levels of Salmonella effectors. We used TNF-α at a concentration of 10 ng/ml, which is similar to the pathologic concentration in an inflamed intestine or patient serum [70]. Using RT-PCR, we investigated the mRNA expression of Salmonella effectors in the pathogenic Salmonella typhimurium SL1344 with or without TNF-α treatment. As shown in Fig. 1A, SipA was up-regulated by TNF-α, whereas gogB and spvB were down-regulated by TNF-α exposure (Fig. 1A). We tested 23 Salmonella effectors, those showing an upregulation of mRNA expression in response to TNF-αare shown in Fig. 1B and those that were downregulated following TNF-α exposure are shown in Fig. 1C. TNF-α significantly upregulated the mRNA expression of SipA, whereas it down-regulated the mRNA expression of gogB and spvB. Overall, this PCR data suggests that certain effectors are responsive to the host inflammatory factor TNF-α.
Levels of effector gene expression determined by RT-PCR in Salmonella SL1344. (A) Representative PCR results for effector mRNA expression. Mdh was used as the internal control. Control: Salmonella without treatment; TNF-α: Salmonella treated with TNF-α. (B) and (C) The relative intensity of PCR bands (Control vs. TNF-α-pretreated Salmonella). There were 3 repeated experiments performed in all controls and TNF-α treated groups. First, the densitometry value of each indicated effector gene was divided by the value for the Mdh band. Next, the obtained values were compared with the corresponding control values. The relative fold changes are shown in Figure 1B and 1C with the control group value set as "1" and compared to the TNF-α treated groups. Data are reported as the mean ± SD of three independent experiments. *P < 0.05 was considered significant.
Responses to TNF-α treatment in Salmonella strains with or without AvrA
Our previous studies found that the Salmonella effector AvrA inhibits the proinflammatory NF-κB pathway (Collier-Hyams et al., 2002) and stabilizes β-catenin and IκBα [71]. We reasoned that the expression levels of AvrA in the bacterial strains may alter their responses to TNF-α treatment. Therefore, we tested effector expression levels in pathogenic Salmonella strains and corresponding AvrA mutants with or without TNF-α treatment. SL14028s with AvrA gene expression is known to express the AvrA protein only at low pH [27]. As shown in Fig. 2, SipA expression was not changed by TNF-α in SL14028s, whereas SipA mRNA in SL1344 was significantly elevated by TNF-α. PhoPC is a mutation derived from SL1344 [63]. Interestingly, the SipA mRNA was undetectable in PhoPC.
The effects of AvrA deletion on effector expression. SipA mRNA expression in the indicated Salmonella strains was determined by PCR. The relative intensity of the PCR bands was analyzed. Control: Salmonella without treatment; TNF-α: Salmonella treated with TNF-α. The data are reported as the mean ± SD of three independent experiments. *P < 0.05 was considered significant.
To confirm that TNF-α pretreatment had no effects on bacterial growth, we measured the optical density (O.D.) of the bacteria in LB after TNF-α treatment for 18 hours. Over the still culture period, no significant difference was observed between the bacterial strain SL1344 with or without TNF-α pretreatment (Fig. 2B). Similar results were found in the SL1344 AvrA-strain with or without TNF-α treatment (Fig. 2C).
In Table 3, we summarize the changes of effector gene expression after TNF-α 18-hour treatment in Salmonella strains with or without AvrA expression.
Alteration of Salmonella effector proteins after TNF-α treatment
Effector protein expression may be different from mRNA levels. We therefore examined strains of Salmonella to determine whether SipA protein levels respond to TNF-α treatment. As shown in Fig. 3, SipA expression was elevated by TNF-α in SL14028s and SL1344. To make sure the difference we observed was not due to protein loading variation, we stained the membrane with Ponceau S Red that indicates total protein levels (Fig. 3B). Relatively equal amounts of proteins in each lane were visible. We also found that SipA and AvrA could not be detected in the AvrA deletion strain derived from SL14028s (Fig. 3A). Without AvrA, SL1344 AvrA-did not alter SipA expression after TNF-α treatment. In addition, we generated an anti-AvrA antibody to detect the level of AvrA protein expression. SL14028s is known to express the AvrA protein only at low pH [26, 27]. Therefore, we did not detect AvrA in the SL14028s group cultured in LB at pH 7.5. AvrA expression is high in the SL1344 strain and increased with TNF-α exposure. Taken together, we found that TNF-α significantly increased SipA protein expression in the pathogenic SL14028s and SL1344 strains (Fig. 3C).
SipA and AvrA protein expression. (A) Western blot assay for the expression of SipA and AvrA. (B) Relative protein band intensity in Ponceau S Red staining. Data are reported as representative results from three independent experiments. (C) The relative intensity of the Western blot bands. The data are reported as the mean ± SD of three independent experiments. *P < 0.05 was considered significant.
TNF-α pretreatment of Salmonella enhances invasion of host cells
We then examined whether pre-treating Salmonella with TNF-α contributes to the physiological function of Salmonella, such as invasion. To determine whether TNF-α contributed to Salmonella invasion, we counted the number of Salmonella invading the human intestinal epithelial HCT116 cells. We found that TNF-α pretreatment of Salmonella increased the amount of internalized bacteria in epithelial cells versus untreated Salmonella SL1344 (Fig. 4A). In the Salmonella SL1344 AvrA-strain, we also found that TNF-α enhanced bacterial invasion of host cells (Fig.4B). Moreover, we examined the number of cell-associated bacteria, including bacteria adhered to and/or internalized into the epithelial monolayers. Our data showed no significant difference of Salmonella associated with epithelial cells with or without TNF-α pretreatment (Fig. 4C SL1344 and Fig. 4D SL1344 AvrA-). Furthermore, we used a JNK inhibitor, SP600125, to treat cells in order to confirm the enhanced bacterial invasion is related to the JNK pathway. Significantly less number of invaded bacteria was found in SL14028S group with SP600125 compared to the no-inhibitor groups (P < 0.05 Fig. 4E). However, invaded bacterial numbers in the TNF pretreatment group and non-TNF treatment group were still significantly different (P < 0.05 Fig. 4D), suggesting that SP600125 could not block the effect of TNF-pretreated Salmonella in enhancing invasion. These in vitro data indicates that TNF-α pretreatment changes the ability of Salmonella to internalize into host cells.
TNF-α pretreatment of Salmonella contributes to enhanced bacterial invasion in human intestinal epithelial HCT116 cells. (A) and (B) Number of internalized Salmonella (A: SL1344; B: SL1344 AvrA-) in the HCT116 cells. (C) and (D) Number of Salmonella associated with HCT116 cells. C: SL1344; D: SL1344 AvrA-. (E) Number of internalized Salmonella in the HCT116 cells with a JNK inhibitor SP600125 (50 μM) pretreatment. HCT116 cells were stimulated with Salmonella with or without TNF-α pretreatment for 30 min, washed, and incubated in fresh DMEM for 30 min. For both cell associated and internalized bacteria, 0.9 ml LB broth was then added and each sample was vigorously mixed and quantitated by plating for CFU on MacConkey agar medium. The mean ± SD is from three replicate experiments.
TNF-α pretreated Salmonella changes the host response
We further hypothesized that TNF-α treatment changes Salmonella effector protein expression, thus altering the host's inflammatory responses. The c-Jun N-terminal kinase (JNK) pathway is known to be regulated by the Salmonella effector AvrA [29, 71]. Salmonella increases JNK phosphorylation [29]. We tested for the alteration of these two pathways as read-outs of inflammatory responses from host cells. We found that TNF-α pretreated Salmonella SL1344 could enhance c-JUN, p-c-JUN, and p-JNK expression in HCT116 cells (Fig. 5A). Statistical data further showed a significant difference in expression of p-c-JUN and p-JNK induced by Salmonella with or without TNF-α treatment (Fig. 5B and 5C). Moreover, we confirm the role of JNK pathway with a JNK inhibitor, SP600125. Inhibitor treatment blocked the enhancement of both p-c-JUN and p-JNK induced by Salmonella with or without TNF-α (Fig. 5D). In addition, we tested the activity of AP-1, a transcription factor which is a heterodimeric protein associated with c-Jun [72]. However, we did not find the difference in induction of AP-1 activity by Salmonella without TNF or with TNF-pretreatment (data not shown).
JNK pathway is activated by S. typhimurium with or without TNF-α pretreatment. A. The expression level of proteins associated with the JNK pathway in intestinal epithelial cells colonized with Salmonella. Intestinal epithelial cells were incubated with an equal number of the indicated S. typhimurium with or without TNF-α pretreatment. Epithelial cells were collected. Cell lysates were immunoblotted with the indicated antibodies. β-actin is an internal control for the Western blot. The data are reported as the mean ± SD of three independent experiments. (B) (C) Relative intensity of Western blot bands. Salmonella exposed to TNF-α induced higher activity of JNK pathway with enhanced p-JNK and p-c-Jun in the host cells. The data are reported as mean ± SD of three independent experiments. *P < 0.05 was considered significant. (D). The expression level of proteins associated with the JNK pathway in intestinal epithelial cells colonized with Salmonella. HCT116 cells were pretreated with a JNK inhibitor SP600125 (50 μM).
IL-8 mRNA and protein levels in intestinal epithelial cells induced by Salmonella with or without TNF-α treatment
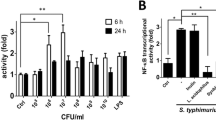
Cytokine IL-8 expression and secretion are common readouts for inflammatory responses in the host cells [73]. It is known that pathogenic Salmonella increases IL-8 through both transcriptional regulation and protein expression levels [58, 71, 73, 74]. We reasoned that exposure to TNF-α makes pathogenic Salmonella more aggressive, inducing more severe inflammatory responses as compared to Salmonella without TNF-α treatment. We assessed the effect of TNF-α exposed Salmonella on IL-8 mRNA expression in human intestinal HCT116 cells. IL-8 mRNA real-time PCR showed that HCT116 cells significantly increased the level of IL-8 mRNA expression after TNF-α pretreated Salmonella colonization (Fig. 6A). In contrast, cells colonized with untreated Salmonella expressed less inflammatory IL-8 mRNA (Fig. 6A). Both pathogenic SL14028s and SL1344 had similar trends: TNF-α pretreated Salmonella induced significantly higher amounts of IL-8 mRNA, over 2.5 folds as compared to untreated Salmonella (Fig. 6A). Furthermore, we examined IL-8 protein secretion into the cell media caused by bacterial infection. As shown in Fig. 6B, an increase in IL-8 protein secretion was detected in the cell media after TNF-α pretreated Salmonella SL14028s colonization for 6 hours. In contrast, less IL-8 protein secretion was induced by untreated Salmonella SL14028s colonization (Fig. 6B). SL1344 had similar trends: TNF-α pretreatment induced significantly higher amounts of IL-8 secretion compared to untreated Salmonella (Fig. 6A). Overall, there is a significant difference of IL-8 secretion in cells colonized with Salmonella strains with or without TNF-α pretreatment. A possibility of the increased IL-8 could be due to the enhanced internalized bacteria after TNF pretreatment. We further tested the relationship between the bacterial loading, intercellular bacterial number and IL-8 secretion. However, we did not find that IL-8 secretion linearly related to the invaded bacterial numbers in the cells (data not shown). The enhanced bacterial invasion by TNF treatment and the increased IL-8 could be two different physiological effects in the host cells. Increased bacterial invasion is not necessary to induce increased IL-8 secretion.
TNF pretreatment of Salmonella contributes to enhanced IL-8 mRNA and proteins in human intestinal epithelial cells. Cells were cultured in DMEM, followed by Salmonella-containing HBSS for 30 min, washed 3 times in HBSS, and incubated at 37°C for 6 hours. Total RNA was extracted for real-time PCR. Cell supernatants were removed and assayed for IL-8 by ELISA. (A) IL-8 mRNA levels in the HCT116 cells after colonization with TNF-pretreated Salmonella. (B) IL-8 protein secreted into the cell culture media in the HCT116 cells after Salmonella infection. (C) IL-8 protein secreted into the cell culture media in the HCT116 cells after Salmonella infection. HCT116 cells were pretreated with a JNK inhibitor SP600125 (50 μM). In a single experiment, samples were assayed in triplicate. The data are reported as mean ± SD of three independent experiments. *P < 0.05 was considered significant.
To confirm the effect of TNF-pretreated Salmonella on IL-8 secretion is through the JNK pathway, we further used the inhibitor SP600125 to treat cells, significant less IL-8 was found in the SL14028S Salmonella with SP600125 group compared to the non-inhibitor group (Fig. 6C P < 0.03). SP600125 treatment was able to decrease the IL-8 secretion significantly in the SL14028 + TNF group v.s. the SP600125 + SL14028 + TNF group (Fig. 6C P = 0.017). There was significant difference in Sl14028S with or without TNF pretreatment (Fig. 6C P < 0.05). However, the difference between TNF pretreatment or no-TNF treatment was abolished after SP600125 pretreatment (Fig. 6C). Taken together, these IL-8 data indicate that TNF-α pretreated Salmonella stimulated more inflammatory responses in the intestinal epithelial cells through the JNK pathway.
Discussion
The aim of this study was to determine the effect of TNF-α on Salmonella effector expression and the ability of TNF-α pretreated Salmonella to induce inflammatory responses in host epithelial cells. We investigated the regulation of Salmonella effectors in a variety of contexts, including mRNA expression, protein expression, and host-bacteria interaction Furthermore, we explored the response of human intestinal cells to TNF-α pretreated Salmonella. Bacterial invasion was enhanced in cells colonized with TNF-α pretreated Salmonella. Salmonella strains with TNF-α pretreatment induced higher proinflammatory responses compared to untreated Salmonella. Overall, our data show that TNF-α exposure makes Salmonella more virulent and enhances inflammation in host intestinal cells. This study provides a new insight into the Salmonella-host interaction in intestinal inflammation and infection.
Our study demonstrates that Salmonella senses the host inflammatory factor TNF-α and responds by changing its effector protein expression and enhancing its virulence, such as invasion. However, it is unknown how Salmonella senses TNF-α in the environment and whether Salmonella has a receptor for TNF-α. Recent findings have begun to reveal the molecular mechanisms by which bacteria can sense small innate immune molecules and modulate virulence gene expression. Wu et al. demonstrated that Pseudomonas aeruginosa recognizes host immune activation and responds by enhancing their virulence phenotype [75]. Also in Pseudomonas, Zaborina et al. showed dynorphin regulation of bacterial pathogenesis and cross-signaling between quorum sensing and quinolone signaling [76]. Norepinephrine modulates interactions between enterohemorrhagic Escherichia coli (EHEC) and the colonic epithelium by increasing bacterial adherence to the colonic mucosa [77]. EHEC uses a QS regulatory system to "sense" that it is within the intestine and then activates genes essential for intestinal colonization [13]. The QS system used by EHEC is known as the LuxS/autoinducer 2 (AI-2) system extensively involved in interspecies communication [13]. Given that eukaryotic cell-to-cell signaling typically occurs through hormones, and bacterial cell-to-cell signaling occurs through QS, QS may be used as a "language" by which bacteria and host cells communicate [13]. In S. typhimurium, the PhoQ sensor kinase is activated by host antimicrobial peptides. PhoQ then promotes the expression of virulence genes through a phosphorelay cascade [78]. However, it is still unknown how pathogenic Salmonella senses TNF-α, thus changing the expression of the bacterial effectors. Studies in Pseudomonas raise the possibility that TNF-α sensors or receptors for TNF-α are encoded for in the prokaryotic genome. Further studies on the Salmonella quorum sensing system and effector regulation will provide insights into this powerful and effective bacteria-host interaction.
Our data demonstrate that TNF-α exposure increases the expression of the Salmonella effector SipA. SipA contributes significantly to Salmonella host cell invasion in vitro and to Salmonella enterocolitis in vivo [35, 41, 79]. SipA also plays key role in maximizing pro-inflammatory responses [41]. Our bacterial invasion study further showed that cells colonized with TNF-α pretreated Salmonella had more internalized Salmonella. Moreover, Salmonella strains with TNF-α pretreatment induced higher proinflammatory responses, such as the activation of JNK and elevation of IL-8, compared to the Salmonella strains without TNF-α exposure. This observation is correlated with the enhanced expression of SipA in Salmonella exposed to TNF-α.
At the mRNA level SL1344 expressing AvrA has a significant ability to modify SipA in response to TNF-α, whereas SL14028s is less responsive (Fig.2A). We also found that an AvrA knockout strain derived from SL14028s had significantly decreased levels of SipA mRNA and protein. Salmonella SL14028s is known to be deficient in AvrA expression. AvrA protein expression was only detectable when SL14028s was cultured in low pH media [25–27]. The status of the effector AvrA may alter the expression of other effectors and the capacity of bacteria to induce host inflammation. Other factors in the environment may also contribute to the expression changes of Salmonella effectors. Although AvrA is known to regulate diverse bacteria-host interactions [28, 29, 71, 80], the synergistic regulation of AvrA and other Salmonella effectors in response to the inflammatory status of the host cells remains unknown. Further investigations on the interaction of AvrA and its fellow effectors will help us to understand the network of Salmonella effectors in epithelial cell-bacteria cross-talk.
TNF-α exposure decreased the mRNA expression of SPI-2 effectors Gog B and SpVB, which are known to promote bacterial replication and systemic spread [19, 20, 22]. We did not examine the protein expression of these two proteins, while the cell culture system limited investigation into the physiologic relevance of the reduction of Gog B and SpVB. Long-term bacterial replication and systematic spread need to be examined in an in vivo model.
In summary, our current study answers the fundamental question of whether TNF-α expressed from host cells can change the expression level of Salmonella effectors, such as SipA, gogB, and spvB. Salmonella exposed to TNF-α induced more bacterial internalization, higher activity of JNK pathway with enhanced p-JNK and p-c-Jun in the host cells. As a consequence of the activation of the JNK pathway, the expression of inflammatory cytokines, such as IL-8, is higher in cells colonized with TNF-α pretreated Salmonella. Overall, Salmonella exposed to TNF-α caused enhanced inflammation in intestinal epithelial cells. We postulate that chronic inflammation with elevated TNF-α in host cells may change the behavior of pathogenic bacterial effectors and may make pathogens more virulent.
Conclusions
We found that TNF-α treatment modulated effector expression in a differential manner. The expression of effector SipA was increased after TNF-α exposure in pathogenic Salmonella. Enhanced bacteria internalization and more severe inflammatory responses of intestinal epithelial cells were found after Salmonella strains were exposed to TNF-α. Activation of the JNK pathway significantly elevates and enhances inflammation in intestinal epithelial cells. As a consequence, the expression of inflammatory cytokines, such as IL-8, is high in cells colonized with TNF-α pretreated Salmonella. Our studies provide new insights into host factor TNF-α regulation of Salmonella effector expression in bacterial invasion and inflammatory responses.
References
Tracey KJ, Cerami A: Tumor necrosis factor: a pleiotropic cytokine and therapeutic target. Annu Rev Med. 1994, 45: 491-503. 10.1146/annurev.med.45.1.491.
Rahman MM, Lucas AR, McFadden G: Viral TNF inhibitors as potential therapeutics. Adv Exp Med Biol. 2009, 666: 64-77. full_text.
Chouaib S, Robinet E, Zyad A, Branellec D: [Tumor necrosis factor: pleiotropic cytokine]. Bull Cancer. 1992, 79: 935-949.
Sidhu RS, Bollon AP: Tumor necrosis factor activities and cancer therapy--a perspective. Pharmacol Ther. 1993, 57: 79-128. 10.1016/0163-7258(93)90037-E.
Keane J: TNF-blocking agents and tuberculosis: new drugs illuminate an old topic. Rheumatology (Oxford). 2005, 44: 714-720. 10.1093/rheumatology/keh567.
Nash PT, Florin TH: Tumour necrosis factor inhibitors. Med J Aust. 2005, 183: 205-208.
Zariffard MR, Novak RM, Lurain N, Sha BE, Graham P, Spear GT: Induction of tumor necrosis factor-alpha secretion and toll-like receptor 2 and 4 mRNA expression by genital mucosal fluids from women with bacterial vaginosis. J Infect Dis. 2005, 191: 1913-1921. 10.1086/429922.
Wang J, Barke RA, Charboneau R, Roy S: Morphine impairs host innate immune response and increases susceptibility to Streptococcus pneumoniae lung infection. J Immunol. 2005, 174: 426-434.
Fejer G, Szalay K, Gyory I, Fejes M, Kusz E, Nedieanu S, Pali T, Schmidt T, Siklodi B, Lazar G: et al., Adenovirus infection dramatically augments lipopolysaccharide-induced TNF production and sensitizes to lethal shock. J Immunol. 2005, 175: 1498-1506.
Turner JD, Langley RS, Johnston KL, Egerton G, Wanji S, Taylor MJ: Wolbachia endosymbiotic bacteria of Brugia malayi mediate macrophage tolerance to TLR- and CD40-specific stimuli in a MyD88/TLR2-dependent manner. J Immunol. 2006, 177: 1240-1249.
Hughes DT, Sperandio V: Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2008, 6: 111-120. 10.1038/nrmicro1836.
Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, Hughson FM: Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell. 2004, 15: 677-687. 10.1016/j.molcel.2004.07.020.
Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB: Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA. 2003, 100: 8951-8956. 10.1073/pnas.1537100100.
Boyen F, Eeckhaut V, Van Immerseel F, Pasmans F, Ducatelle R, Haesebrouck F: Quorum sensing in veterinary pathogens: mechanisms, clinical importance and future perspectives. Vet Microbiol. 2009, 135: 187-195. 10.1016/j.vetmic.2008.12.025.
Mota LJ, Cornelis GR: The bacterial injection kit: type III secretion systems. Ann Med. 2005, 37: 234-249. 10.1080/07853890510037329.
Coburn B, Sekirov I, Finlay BB: Type III secretion systems and disease. Clin Microbiol Rev. 2007, 20: 535-549. 10.1128/CMR.00013-07.
Coburn B, Li Y, Owen D, Vallance BA, Finlay BB: Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect Immun. 2005, 73: 3219-3227. 10.1128/IAI.73.6.3219-3227.2005.
Mattoo S, Lee YM, Dixon JE: Interactions of bacterial effector proteins with host proteins. Curr Opin Immunol. 2007, 19: 392-401. 10.1016/j.coi.2007.06.005.
McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V: Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol. 2009, 12: 117-124. 10.1016/j.mib.2008.12.001.
Coombes BK, Wickham ME, Brown NF, Lemire S, Bossi L, Hsiao WW, Brinkman FS, Finlay BB: Genetic and molecular analysis of GogB, a phage-encoded type III-secreted substrate in Salmonella enterica serovar typhimurium with autonomous expression from its associated phage. J Mol Biol. 2005, 348: 817-830. 10.1016/j.jmb.2005.03.024.
Xu X, Hensel M: Systematic analysis of the SsrAB virulon of Salmonella enterica. Infect Immun. 78: 49-58. 10.1128/IAI.00931-09.
Browne SH, Hasegawa P, Okamoto S, Fierer J, Guiney DG: Identification of Salmonella SPI-2 secretion system components required for SpvB-mediated cytotoxicity in macrophages and virulence in mice. FEMS Immunol Med Microbiol. 2008, 52: 194-201. 10.1111/j.1574-695X.2007.00364.x.
Raffatellu M, Wilson RP, Chessa D, Andrews-Polymenis H, Tran QT, Lawhon S, Khare S, Adams LG, Baumler AJ: SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype typhimurium invasion of epithelial cells. Infect Immun. 2005, 73: 146-154. 10.1128/IAI.73.1.146-154.2005.
Schlumberger MC, Kappeli R, Wetter M, Muller AJ, Misselwitz B, Dilling S, Kremer M, Hardt WD: Two newly identified SipA domains (F1, F2) steer effector protein localization and contribute to Salmonella host cell manipulation. Mol Microbiol. 2007, 65: 741-760. 10.1111/j.1365-2958.2007.05823.x.
Hardt WD, Galan JE: A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc Natl Acad Sci USA. 1997, 94: 9887-9892. 10.1073/pnas.94.18.9887.
Ben-Barak Z, Streckel W, Yaron S, Cohen S, Prager R, Tschape H: The expression of the virulence-associated effector protein gene avrA is dependent on a Salmonella enterica-specific regulatory function. Int J Med Microbiol. 2006, 296: 25-38. 10.1016/j.ijmm.2005.08.004.
Streckel W, Wolff AC, Prager R, Tietze E, Tschape H: Expression profiles of effector proteins SopB, SopD1, SopE1, and AvrA differ with systemic enteric, and epidemic strains of Salmonella enterica. Mol Nutr Food Res. 2004, 48: 496-503. 10.1002/mnfr.200400035.
Du F, Galan JE: Selective inhibition of type III secretion activated signaling by the Salmonella effector AvrA. PLoS Pathog. 2009, 5: e1000595-10.1371/journal.ppat.1000595.
Jones RM, Wu H, Wentworth C, Luo L, Collier-Hyams L, Neish AS: Salmonella AvrA Coordinates Suppression of Host Immune and Apoptotic Defenses via JNK Pathway Blockade. Cell Host Microbe. 2008, 3: 233-244. 10.1016/j.chom.2008.02.016.
Liao AP, Petrof EO, Kuppireddi S, Zhao Y, Xia Y, Claud EC, Sun J: Salmonella type III effector AvrA stabilizes cell tight junctions to inhibit inflammation in intestinal epithelial cells. PLoS ONE. 2008, 3: e2369-10.1371/journal.pone.0002369.
Lu Y, Ye L, Yu S, Zhang S, Xie Y, McKee MD, Li YC, Kong J, Eick JD, Dallas SL, Feng JQ: Rescue of odontogenesis in Dmp1-deficient mice by targeted re-expression of DMP1 reveals roles for DMP1 in early odontogenesis and dentin apposition in vivo. Dev Biol. 2007, 303: 191-201. 10.1016/j.ydbio.2006.11.001.
Liu X, Lu R, Wu S, Sun J: Salmonella regulation of intestinal stem cells through the Wnt/beta-catenin pathway. FEBS Lett.
Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J: et al., Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007, 5: 2177-2189. 10.1371/journal.pbio.0050244.
Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH, Galan JE: Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog. 2009, 5: e1000538-10.1371/journal.ppat.1000538.
Hapfelmeier S, Ehrbar K, Stecher B, Barthel M, Kremer M, Hardt WD: Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun. 2004, 72: 795-809. 10.1128/IAI.72.2.795-809.2004.
Jones MA, Wood MW, Mullan PB, Watson PR, Wallis TS, Galyov EE: Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect Immun. 1998, 66: 5799-5804.
Galyov EE, Wood MW, Rosqvist R, Mullan PB, Watson PR, Hedges S, Wallis TS: A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol. 1997, 25: 903-912. 10.1111/j.1365-2958.1997.mmi525.x.
Ehrbar K, Friebel A, Miller SI, Hardt WD: Role of the Salmonella pathogenicity island 1 (SPI-1) protein InvB in type III secretion of SopE and SopE2, two Salmonella effector proteins encoded outside of SPI-1. J Bacteriol. 2003, 185: 6950-6967. 10.1128/JB.185.23.6950-6967.2003.
Zhou D, Galan J: Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 2001, 3: 1293-1298. 10.1016/S1286-4579(01)01489-7.
Bakowski MA, Braun V, Brumell JH: Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic. 2008, 9: 2022-2031. 10.1111/j.1600-0854.2008.00827.x.
Wall DM, Nadeau WJ, Pazos MA, Shi HN, Galyov EE, McCormick BA: Identification of the Salmonella enterica serotype typhimurium SipA domain responsible for inducing neutrophil recruitment across the intestinal epithelium. Cell Microbiol. 2007, 9: 2299-2313. 10.1111/j.1462-5822.2007.00960.x.
Le Negrate G, Faustin B, Welsh K, Loeffler M, Krajewska M, Hasegawa P, Mukherjee S, Orth K, Krajewski S, Godzik A: et al., Salmonella secreted factor L deubiquitinase of Salmonella typhimurium inhibits NF-kappaB, suppresses IkappaBalpha ubiquitination and modulates innate immune responses. J Immunol. 2008, 180: 5045-5056.
Rytkonen A, Poh J, Garmendia J, Boyle C, Thompson A, Liu M, Freemont P, Hinton JC, Holden DW: SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci USA. 2007, 104: 3502-3507. 10.1073/pnas.0610095104.
Coombes BK, Lowden MJ, Bishop JL, Wickham ME, Brown NF, Duong N, Osborne S, Gal-Mor O, Finlay BB: SseL is a salmonella-specific translocated effector integrated into the SsrB-controlled salmonella pathogenicity island 2 type III secretion system. Infect Immun. 2007, 75: 574-580. 10.1128/IAI.00985-06.
Haraga A, Miller SI: A Salmonella type III secretion effector interacts with the mammalian serine/threonine protein kinase PKN1. Cell Microbiol. 2006, 8: 837-846. 10.1111/j.1462-5822.2005.00670.x.
Wu S, Ye Z, Liu X, Zhao Y, Xia Y, Steiner A, Petrof EO, Claud EC, Sun J: Salmonella typhimurium infection increases p53 acetylation in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2010, 298: G784-94. 10.1152/ajpgi.00526.2009.
Sun J: Pathogenic Bacterial Proteins and their Anti-Inflammatory Effects in the Eukaryotic Host. Antiinflamm Antiallergy Agents Med Chem. 2009, 8: 214-227.
Troyanovsky SM: Mechanism of cell-cell adhesion complex assembly. Curr Opin Cell Biol. 1999, 11: 561-566. 10.1016/S0955-0674(99)00021-6.
Darwin KH, Miller VL: Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin Microbiol Rev. 1999, 12: 405-428.
McCormick BA, Miller SI, Carnes D, Madara JL: Transepithelial signaling to neutrophils by salmonellae: a novel virulence mechanism for gastroenteritis. Infect Immun. 1995, 63: 2302-2309.
Takeuchi A: Electron microscope studies of experimental Salmonella infection. I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am J Pathol. 1967, 50: 109-136.
Galan JE, Pace J, Hayman MJ: Involvement of the epidermal growth factor receptor in the invasion of cultured mammalian cells by Salmonella typhimurium. Nature. 1992, 357: 588-589. 10.1038/357588a0.
McCormick BA, Colgan SP, Delp-Archer C, Miller SI, Madara JL: Salmonella typhimurium attachment to human intestinal epithelial monolayers: transcellular signalling to subepithelial neutrophils. J Cell Biol. 1993, 123: 895-907. 10.1083/jcb.123.4.895.
Gewirtz AT, Rao AS, Simon PO, Merlin D, Carnes D, Madara JL, Neish AS: Salmonella typhimurium induces epithelial IL-8 expression via Ca(2 +)-mediated activation of the NF-kappaB pathway. J Clin Invest. 2000, 105: 79-92. 10.1172/JCI8066.
Finlay BB, Falkow S: Salmonella interactions with polarized human intestinal Caco-2 epithelial cells. J Infect Dis. 1990, 162: 1096-1106.
Francis CL, Starnbach MN, Falkow S: Morphological and cytoskeletal changes in epithelial cells occur immediately upon interaction with Salmonella typhimurium grown under low-oxygen conditions. Mol Microbiol. 1992, 6: 3077-3087. 10.1111/j.1365-2958.1992.tb01765.x.
Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD: Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003, 71: 2839-2858. 10.1128/IAI.71.5.2839-2858.2003.
Duan Y, Liao AP, Kuppireddi S, Ye Z, Ciancio MJ, Sun J: beta-Catenin activity negatively regulates bacteria-induced inflammation. Lab Invest. 2007, 87: 613-624. 10.1038/labinvest.3700497.
Kohler H, Sakaguchi T, Hurley BP, Kase BA, Reinecker HC, McCormick BA: Salmonella enterica serovar Typhimurium regulates intercellular junction proteins and facilitates transepithelial neutrophil and bacterial passage. Am J Physiol Gastrointest Liver Physiol. 2007, 293: G178-187. 10.1152/ajpgi.00535.2006.
Martinez-Argudo I, Jepson MA: Salmonella translocates across an in vitro M cell model independently of SPI-1 and SPI-2. Microbiology. 2008, 154: 3887-3894. 10.1099/mic.0.2008/021162-0.
Gradel KO, Nielsen HL, Schonheyder HC, Ejlertsen T, Kristensen B, Nielsen H: Increased short- and long-term risk of inflammatory bowel disease after salmonella or campylobacter gastroenteritis. Gastroenterology. 2009, 137: 495-501. 10.1053/j.gastro.2009.04.001.
Andoh A, Yagi Y, Shioya M, Nishida A, Tsujikawa T, Fujiyama Y: Mucosal cytokine network in inflammatory bowel disease. World J Gastroenterol. 2008, 14: 5154-5161. 10.3748/wjg.14.5154.
Miller SI, Mekalanos JJ: Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol. 1990, 172: 2485-2490.
Hamilton CM, Aldea M, Washburn BK, Babitzke P, Kushner SR: New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989, 171: 4617-4622.
Miller VL, Mekalanos JJ: A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988, 170: 2575-2583.
Boyd EF, Nelson K, Wang FS, Whittam TS, Selander RK: Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci USA. 1994, 91: 1280-1284. 10.1073/pnas.91.4.1280.
Lissner CR, Swanson RN, O'Brien AD: Genetic control of the innate resistance of mice to Salmonella typhimurium: expression of the Ity gene in peritoneal and splenic macrophages isolated in vitro. J Immunol. 1983, 131: 3006-3013.
Sun J, Hobert ME, Rao AS, Neish AS, Madara JL: Bacterial activation of beta-catenin signaling in human epithelia. Am J Physiol Gastrointest Liver Physiol. 2004, 287: G220-227. 10.1152/ajpgi.00498.2003.
Sun J, Kong J, Duan Y, Szeto FL, Liao A, Madara JL, Li YC: Increased NF-kappaB activity in fibroblasts lacking the vitamin D receptor. Am J Physiol Endocrinol Metab. 2006, 291: E315-322. 10.1152/ajpendo.00590.2005.
Komatsu M, Kobayashi D, Saito K, Furuya D, Yagihashi A, Araake H, Tsuji N, Sakamaki S, Niitsu Y, Watanabe N: Tumor necrosis factor-alpha in serum of patients with inflammatory bowel disease as measured by a highly sensitive immuno-PCR. Clin Chem. 2001, 47: 1297-1301.
Ye Z, Petrof EO, Boone D, Claud EC, Sun J: Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol. 2007, 171: 882-892. 10.2353/ajpath.2007.070220.
Hess J, Angel P, Schorpp-Kistner M: AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004, 117: 5965-5973. 10.1242/jcs.01589.
Neish AS, Gewirtz AT, Zeng H, Young AN, Hobert ME, Karmali V, Rao AS, Madara JL: Prokaryotic regulation of epithelial responses by inhibition of IkappaB-alpha ubiquitination. Science. 2000, 289: 1560-1563. 10.1126/science.289.5484.1560.
Gewirtz AT, Simon PO, Schmitt CK, Taylor LJ, Hagedorn CH, O'Brien AD, Neish AS, Madara JL: Salmonella typhimurium translocates flagellin across intestinal epithelia, inducing a proinflammatory response. J Clin Invest. 2001, 107: 99-109. 10.1172/JCI10501.
Wu L, Estrada O, Zaborina O, Bains M, Shen L, Kohler JE, Patel N, Musch MW, Chang EB, Fu YX: et al., Recognition of host immune activation by Pseudomonas aeruginosa. Science. 2005, 309: 774-777. 10.1126/science.1112422.
Zaborina O, Lepine F, Xiao G, Valuckaite V, Chen Y, Li T, Ciancio M, Zaborin A, Petrof EO, Turner JR: et al., Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog. 2007, 3: e35-10.1371/journal.ppat.0030035.
Green BT, Lyte M, Chen C, Xie Y, Casey MA, Kulkarni-Narla A, Vulchanova L, Brown DR: Adrenergic modulation of Escherichia coli O157:H7 adherence to the colonic mucosa. Am J Physiol Gastrointest Liver Physiol. 2004, 287: G1238-1246. 10.1152/ajpgi.00471.2003.
Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI: Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005, 122: 461-472. 10.1016/j.cell.2005.05.030.
Zhang S, Santos RL, Tsolis RM, Stender S, Hardt WD, Baumler AJ, Adams LG: The Salmonella enterica serotype typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect Immun. 2002, 70: 3843-3855. 10.1128/IAI.70.7.3843-3855.2002.
Collier-Hyams LS, Zeng H, Sun J, Tomlinson AD, Bao ZQ, Chen H, Madara JL, Orth K, Neish AS: Cutting edge: Salmonella AvrA effector inhibits the key proinflammatory, anti-apoptotic NF-kappa B pathway. J Immunol. 2002, 169: 2846-2850.
Acknowledgements
We thank members of the Sun laboratory for their technical support and helpful discussion, Dr. Michelle Dziejman for helping with the AvrA knockout in SL14028s, and Drs. Chang-Ho Baek and Roy Curtiss III (Arizona State University) for providing the anti-SipA antibody. We also thank Yuxuan Xia for critical editing of this manuscript.
This work was supported by NIDDK KO1 DK075386, NIDDK R03DK089010-01, and the American Cancer Society RSG-09-075-01-MBC awarded to Jun Sun. There are no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
All authors read and approved the final manuscript.
JM: Participated in the experimental design, preparation of the RNA sample, PCR analysis, Western Blots, ELISA, acquisition of data, analysis and interpretation of data, and drafting of tables and figures. JM also helped to write Methods for the manuscript.
YZ: Participated in growing bacteria, bacterial invasion assays, Western Blots, ELISA, JNK inhibitor treatment, AvrA mutant strain establishment, interpretation of the data, statistical analysis, analysis and interpretation of data, and drafting of tables and figures.
YX: Performed statistical analysis and made critical revision of the manuscript for intellectual content.
JS: Participated in the study concept and design, acquisition of data, analysis and interpretation of data, material support, drafting and critical revision of the manuscript for important intellectual content. JS also obtained funding and supervised the study.
Jun Ma, Yong-guo Zhang contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ma, J., Zhang, Yg., Xia, Y. et al. The inflammatory cytokine tumor necrosis factor modulates the expression of Salmonella typhimurium effector proteins. J Inflamm 7, 42 (2010). https://doi.org/10.1186/1476-9255-7-42
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-9255-7-42