Abstract
Background
Posttranslational protein modifications are known to modulate key biological processes like proliferation and apoptosis. Accumulating evidence shows that ST6GAL1, an enzyme that catalyzes the transfer of sialic acid onto galactose-containing substrates, is aberrantly expressed in various cancers and may affect cell motility and invasion. This is the first study to describe ST6GAL1 expression and regulation in human bladder cancer.
Methods
ST6GAL1 mRNA expression levels in human cell lines (UROtsa, RT4, RT112 and J82) and tissue samples (n = 15 normal urothelium (NU), n = 13 papillary non-invasive tumors (pTa), n = 12 carcinoma in situ (CIS), n = 26 muscle invasive tumors (pT2-4)) were assessed using real-time PCR. In addition, ST6GAL1 protein expression was evaluated using immunohistochemistry. Promoter methylation analysis was performed using methylation-specific PCR (MSP) in cell lines (n = 4) and patient samples (n = 23 NU, n = 12 CIS, n = 29 pTa, n = 41 pT2-4). Epigenetic ST6GAL1 gene silencing was confirmed by in vitro demethylation of bladder cell lines. Data were validated by analysis of an independent bladder tumor data set (n = 184) based on The Cancer Genome Atlas (TCGA) portal.
Results
Semi-quantitative ST6GAL1 real-time PCR expression analysis showed two distinct trends: In muscle-invasive tumors ST6GAL1 expression was downregulation by 2.7-fold, while papillary non-invasive tumors showed an increased ST6GAL1 mRNA expression compared to normal urothelium. ST6GAL1 loss in muscle-invasive tumors was associated with increasing invasiveness. On the protein level, 69.2% (n = 45/65) of all tumors showed a weak ST6GAL1 protein staining (IRS ≤ 4) while 25.6% (16/65) exhibited a complete loss (IRS = 0) of ST6GAL1 protein. Tumor-specific DNA methylation of the ST6GAL1 promoter region was frequently found in pT2-4 tumors (53.6% (22/41)), whereas only 13.8% (4/29) of pTa tumors showed ST6GAL1 promoter methylation. Normal urothelium remained unmethylated. Importantly, we significantly revealed an inverse correlation between ST6GAL1 mRNA expression and ST6GAL1 promoter merthylation in primary bladder cancer. These findings were clearly verified by the TCGA public data set and in vitro demethylation assays functionally confirmed ST6GAL1 promoter methylation as a potential regulatory factor for ST6GAL1 gene silencing.
Conclusions
Our study characterizes for the first time ST6GAL1 expression loss caused by aberrant ST6GAL1 promoter methylation potentially indicating a tumor suppressive role in bladder carcinogenesis.
Similar content being viewed by others
Background
Urinary bladder cancer currently represents the 5th most common cancer type in the industrialized nations [1]. Clinically, bladder cancer poses a unique clinical challenge, consisting of a heterogeneous group with either recurrent, but relatively benign course or progressive oncological course [2]. In 90% of cases, tumors arise from superficial cell layers in the urogenital tract known as urothelial cells (formerly transitional cells) [3]. Here, two major growth forms, papillary non-invasive and flat-invasive, have been identified, underlying two separate molecular pathways characterized by distinct mutations [4]. Low-grade tumors, which display a papillary growth form and are mostly superficial (Ta low grade urothelial carcinoma (UC)), constitute the largest group at diagnosis, and are characterized by their high recurrence rate. The second group is formed by high-grade tumors and includes carcinoma in situ (CIS), a flat high-grade lesion, which in 60–80% of cases progresses to invasive bladder cancer within 5 years [3]. Low stage/grade tumors are often characterized by mutations in fibroblast growth factor receptor 3 (FGFR3)[5, 6] and rat sarcoma (RAS) genes [5, 7], while flat carcinoma in situ lesions frequently show mutations in TP53 in addition to a loss of heterozygosity of the retinoblastoma (RB) gene [3, 5, 8, 9]. As such high grade tumors progress to a muscle invasive stage, an ever increasing degree of DNA hypermethylation is observed [10]. Current research aims to further elucidate the changes defining these divergent growth forms given their different clinical impact and therapeutic needs.
Post-translational modifications are known to influence protein characteristics such as protein folding and stability, thus modulating biological processes like cell growth and migration [11]. As a result, altered glycosylation of proteins, such as cell surface glycoproteins and glycolipids, is a common and frequent feature during carcinogenesis, due to impaired activity of glycosyltransferases [12]. The ST6GAL1 gene encodes a type II membrane protein (beta-galactosamide alpha-2,6-sialyltransferase 1, ST6GAL1) that catalyzes the transfer of sialic acid from cytidine-monophosphate (CMP)-sialic acid onto galactose-containing substrates [12–14]. Previous studies have shown that ST6GAL1 functions as a critical regulator of cell survival in several cell death pathways [14, 15]. For example, its sialylation of the Fas death receptor hinders internalization of Fas after activation, thereby reducing apoptotic signaling [14]. Similarly, ST6GAL1 promotes cell surface retention of the tumor necrosis factor receptor 1 (TNFR1) and the CD45 receptor [14, 15]. Furthermore in vitro studies have shown that ST6GAL1 upregulation promotes cell migration and invasion through its interaction with the B1 integrin receptor [16–19], while animal models of colon cancer implicate ST6GAL1 in tumor invasiveness [20]. While these studies clearly underline the oncogenic potential of ST6GAL1, seemingly contradictory evidence has emerged suggesting it may also have tumor suppressive qualities [21]. For example, recent studies clearly showed that a downregulation of ST6GAL1 activity in colorectal carcinoma cell lines facilitated cell proliferation and tumor growth [21].
However, the impact of ST6GAL1 in bladder cancer remains unclear to date. We found ST6GAL1 downregulated in bladder cancer samples in a previous metg001A Affymetrix® GeneChip study reported by Wild et al. [22]. Therefore, the current study seeks to further elucidate ST6GAL1 expression and its regulation in order to determine the potential impact of the glycosyltransferase ST6GAL1 on bladder cancer development.
Methods
Urothelial cell lines
The human SV40-transfected urothelial cell line UROtsa, initially generated from normal ureter tissue, and the papillary-invasive urinary bladder cancer cell lines RT4 and RT112, as well as the invasive bladder cancer cell line J82 from ATCC (American Type Culture Collection, Manassas, Virginia, USA) were cultivated according to the manufacturer’s instructions.
Patient materials
Formalin fixed paraffin embedded (FFPE) tumorous bladder tissue samples analyzed in this study were obtained from our pathology archive and the “whole bladder sampling project (bladder mapping)” integrated in the tumor bank of the Euregional comprehensive Cancer Center Aachen (ECCA), now part of the RWTH centralized biomaterial bank (RWTH cBMB; http://www.cbmb.rwth-aachen.de). All cBMB patients gave written informed consent for retention and analysis of their tissue for research purposes according to local Institutional Review Board (IRB)-approved protocols (approval no. EK-206/09, EK-122/04 and EK 173/06) of the medical faculty of the RWTH Aachen University. For cohort characteristics of all analyzed samples see Table 1. Additionally, 15 normal tissue samples of patients were used. In order to prevent contamination from surrounding tissue, urothelium or tumor tissue was isolated from multiple tissue sections via manual micro-dissection under a stereo microscope, respectively.
DNA and RNA extraction
DNA extraction was achieved using the QiAmp-DNA-Mini-Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. RNA was isolated using the TRIzol approach specified by the manufacturers (Invitrogen, Carlsbad/CA, USA).
Reverse Transcription PCR (RT-PCR)
A total of 1 μg RNA was translated into cDNA using the Promega-Reverse-Transcription-System (Promega, Madison/WI, USA), according to the manufacturer’s instructions In order to maximize the cDNA yield, Oligo-dTs and hexameric random primers (pdN(6)) were mixed in a ratio of 1:2. A total of 1 μl cDNA (20 ng) was used for the PCR reaction.
Semi-quantitative real-time PCR
Real-time PCRs were performed using the iCycler system iQ5 (Bio-Rad Laboratories, Munich) with an intron bridging primer, according to the manufacturer’s instructions. The ubiquitous housekeeping gene glyeradehyde 3-phosphate dehydrogenase (GAPDH) was used as a reference gene. For analysis, a cut off value for GAPDH was assigned at a value of 30. The sequences of the applied primers were as follows: GAPDH 5′-sense: 5′-GAA GGT GAA GGT CGG AGT CA-3′; 3′-antisense: 5′-AAT GAA GGG CTC ATT GAT GG-3′ with a product size of 108bp. ST6GAL1 5′-sense: 5′-TGT CTA GAA AAG AAG GTG GAG ACA T-3′; 3′-antisense: 5′-AGG GTC CTG TTG GCA TTC TC-3′ with a product size of 89 bp. The annealing temperature for both primer sets was 60°C. The relative mRNA-quantification was analyzed using comparative CT methods in comparison to GAPDH-expression.
Bisulfite-modification and methylation-specific PCR (MSP)
Bisulfite conversion of 1 μg of all genomic DNA was achieved using the EZ-DNA-Methylation-Kit (Zymo Research, Orange/CA, USA) and the precipitate was eluted in 20 μl of Tris-Ethylenediaminetetraacetic acid (EDTA)-buffer. Thereafter, 1 μl was amplified in a methylation specific PCR using an optimized PCR buffer. MSP-Primers, specific for the unmethylated ST6GAL1-promotor sequence, were used and read as follows: 5′-GAA GAC GTT TGG GGT ATT GTT CGG C-3′ (M sense) and 5′-TAC ACT CTC GAC CGC GAA AAC TAC G-3′ (M antisense); 5′-GGG AAG ATG TTT GGG GTA TTG TTT GGT G-3′ (U sense) and 5′-TCA CTC ACT ACA CTC TCA ACC GCA AAA ACT ACA-3′ (U antisense). The reaction consisted of 400 nM of the specific primer pairs and 1.25 mM of individual dNTPs. The PCR was completed using the hotstart-PCR method, where 1.25 units of Taq-DNA-Polymerase (Roche Diagnostics, Mannheim, Germany) were given to the mixture when the reaction reached a temperature of 80°C. The PCR conditions were: 95°C for 5 min, followed by 35 cycles of the following sequence: 95°C for 30 s, 56°C for 30 s, 72°C for 30 s and 72°C for 5 min. The amplified PCR product was then run on a 3% low-range ultra-agarose gel (Bio-Rad Laboratories, Hercules/CA, Germany) with ethidium bromide and then visualized using ultraviolet light.
In vitrodemethylation of genomic DNA
Bladder cell lines were seeded in a 6 well dish with a concentration of 3 × 104 cells/cm. A demethylating substance, 5-aza-2′-deoxycytidine (DAC) in a concentration of 1 μM (Sigma-Aldrich, Deisenheim, Germany), was added with fresh medium on day 1, 2 und 3. In addition, 300 nM of the histone deacetylase inhibitor trichostatin A (TSA, Sigma-Aldrich) was added on day 3. The cells were cultivated in fresh medium after every treatment and harvested on the fourth day for RNA extraction.
ST6GAL1 immunohistochemistry
2 μm slides of formalin fixed paraffin embedded (FFPE) bladder tissues were stained with ST6GAL1 monoclonal antibody LN1 clone with 1:100 dilution (MAB6959, Abnova, Walnut, CA, USA). Heat induced antigen retrieval in pH 9.0 EDTA buffer was performed and samples were blocked with peroxide blocking solution from DAKO (DAKO, Hamburg, Germany). DAKO K5007 kit was used as a detection method, according to the manufacturer’s instructions. For visualization 3,3′-diaminobenzidine (DAB) and haematoxylin counter-stain was used. Staining was evaluated according to an adapted semi quantitative scoring system by Remmele and Stegner [23].
Validation of ST6GAL1expression and promoter hypermethylation in an independent set of bladder tumors
In order to independently assess ST6GAL1 mRNA expression and DNA methylation we used public data from both primary invasive [24] and papillary bladder cancer tissues from “The Cancer Genome Atlas” (TCGA) data portal (https://tcga-data.nci.nih.gov). These comprise data of overall n = 184 patients from two independent platforms: Illumina Infinium DNA methylation chip (HumanMethylation 450) and Illumina HiSeq gene expression. The data can be explored through the cBio Cancer Genomics Portal (http://cbioportal.org). For cohort characteristics of analyzed TCGA samples in this current study see Additional file 1: Table S1.
Statistical data analysis
All statistical analysis was done using SPSS 17.0 (SPSS Software GmbH, Munich). Results were considered to be statistically significant given a p value of <0.05. All statistical tests were performed 2 sided. In order to compare two groups, the non-parametric Mann–Whitney U-test was implemented. In case of more than two generated groups the Kruskal-Wallis test and Dunn’s multiple comparison test was used. Any correlation to clinical pathological factors and molecular parameters were analyzed using a descriptive Fisher’s exact-test. Correlation between ST6GAL1 expression (TCGA Illumina HiSeq platform) and ST6GAL1 methylation data (TCGA HM450 platform) was performed by calculating a Spearman correlation coefficient.
Results
ST6GAL1mRNA is differently expressed in human bladder cancer
Previously, Wild and colleagues described novel candidate genes differentially expressed in human bladder cancer [22]. Based on that, we re-analyzed the DNA microarray data set by using in silico database mining procedures identifying novel candidates with differential expression in the course of bladder cancer progression (data not shown). As such we frequently detected ST6GAL1 expression loss by 2.6-fold in pTa and by 3.4-fold in pT1/pT2 bladder tumors. Therefore we aimed to study for the first time ST6GAL1 expression and regulation in bladder cancer.
Semi-quantitative ST6GAL1 real-time PCR expression analysis was performed on 51 bladder cancer tissue samples including flat (n = 12 CIS), papillary non-invasive (n = 13 pTa), and muscle-invasive (n=26 pT2-4) bladder tumor samples (Figure 1A). Detailed cohort statistics can be found in Table 1. Overall, no significant trend was observed when normal urothelium was compared with a mixed tumor cohort (1B). Classification of these samples by tumor subtypes, however showed that ST6GAL1 mRNA expression tends to be downregulated (fold change, FC: 0.36) in muscle-invasive tumors when compared to normal urothelium (NU) (n = 15). In contrast, ST6GAL1 mRNA expression in papillary non-invasive tumors was found to be increased (FC: 1.88) (Figure 1C). A tight association between loss of ST6GAL1 mRNA expression and advanced bladder tumor stages (pT2-pT4; invasive subtype) was significantly underscored by using Fisher’s exact test (Table 2).
ST6GAL1 mRNA expression analyses in human bladder cancer. (A) Real-time PCR based ST6GAL1 mRNA expression analyses of 51 tumor samples (UC) compared to normal urothelium (NU) samples (n = 15). The median expression level of NU was set to 1. Vertical lines: ± standard error of margin (s.e.m.). (B) Box plot showing median ST6GAL1 mRNA expression in normal urothelium compared to all analyzed urothelial cancer samples. Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak and minimum, ns: not significant. (C) Itemized box plot demonstrating median ST6GAL1 mRNA expression in normal urothelium (NU), non-invasive papillary tumors (PAP), CIS, and invasive bladder tumors (INV). Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak and minimum. (D) Real-time PCR based ST6GAL1 mRNA expression analyses of patient triplets with matched normal urothelium, CIS and invasive tumor samples. Matched CIS and solid tumors were normalized to the corresponding NU, respectively. Vertical lines: + s.e.m.
Facing this heterogeneous expression in bladder tumors, we performed further real-time PCR expression analysis using eight patient-matched specimens including NU, CIS, and invasive tumor tissues from each single patient allowing a precise assessment of ST6GAL1 mRNA expression in the course of bladder cancer progression (Figure 1D). In five of the analyzed patients, a clear downregulation (<0.5 FC) of ST6GAL1 mRNA expression was detected in solid tumors, i.e. in invasive stages. Only one patient showed an increase with a FC >2 in ST6GAL1 gene expression and two patients an insignificant change (Figure 1D), not impairing a pronounced loss of ST6GAL1 in invasive tumor stages.
ST6GAL1 protein expression in human bladder cancer
In light of differential ST6GAL1 mRNA expression in bladder cancer, we performed immunohistochemical ST6GAL1 protein expression in NU as well as in bladder tumor tissues. ST6GAL1 protein staining was quantified according to an adapted immunoreactive score (IRS) developed by Remmele and Stegner [23]. In NU ST6GAL1 was detected cytoplasmatically in all urothelial cell layers, with strongest abundance in superficial (“umbrella”) cells (Figure 2A). In contrast, invasive bladder tumors were characterized by a decreased ST6GAL1 protein staining (Figures 2B-D): 69.2% (n = 45/65) of all analyzed tumors showed an ST6GAL1 protein staining of IRS ≤ 4 and 25.6% (16/65) exhibited an almost complete loss (IRS = 0) of ST6GAL1 protein (Figure 2G). A correlation analysis with clinico-pathological characteristics revealed a significant association (p = 0.017) of ST6GAL1 protein reduction and the existence of lymph node metastasis (Table 3).
ST6GAL1 protein expression in human bladder cancer. (A) + (B) matched samples of patient #1 with (A) normal urothelium with accentuated ST6GAL1 protein expression in superficial (“umbrella”) cells and (B) complete loss of ST6GAL1 protein in the poorly differentiated invasive bladder cancer. (C) + (D) Matched samples of patient #2 with (C) normal urothelium with accentuated ST6GAL1 protein expression in superficial (“umbrella”) cells and (D) moderate ST6GAL1 protein expression in the invasive bladder cancer (staining intensity 2). (E) Positive control: prostate sample with strong ST6GAL1 staining (staining intensity 3). (F) Negative control: prostate sample with no staining (staining intensity 0). (G) Histogram of ST6GAL1 protein expression (immunoreactive score, IRS) distribution among all tumor samples.
ST6GAL1promoter hypermethylation is associated with ST6GAL1 expression loss in human bladder cell lines
Given the observation of ST6GAL1 expression loss in the course of bladder cancer progression, we attempted to decipher the molecular cause for ST6GAL1 gene silencing. Recently, DNA methylation in promoter regions, a frequent and well-known epigenetic mechanism of gene inactivation during carcinogenesis was shown in breast cancer [25]. Therefore, we assessed whether this epigenetic modification might be responsible for ST6GAL1 downregulation in bladder cancer as well. Analysis of the ST6GAL1 gene promoter using the genomic DNA information (ENSEMBL contig ENSG00000073849) and the Methprimer program [26] identified two CpG-rich islands between genomic positions 186,930,485 and 187,078,553 on chromosome 3 which met the following criteria: DNA region: ≥200bp; Obs/Exp: ≥0.6; %GC: ≥50. The ST6GAL1 promoter region analyzed by methylation specific PCR (MSP) is located in the non-coding exon 1 next to the transcription start site (TSS) and encodes potential regulatory sequences such as the ubiquitous transcription factor II B (TFIIB) recognition element (BRE: ccgCGCC ) (Figure 3A).
ST6GAL1 promoter methylation in human bladder cell lines correlates with ST6GAL1 mRNA expression. (A) Schematic map of the human ST6GAL1 promoter region including the relative positions of analyzed CpG dinucleotides using MSP (MSP primer binding sites are indicated by arrows) that is located within the non-coding exon 1 region. Two predicted CpG islands are located between base -442 and base +136 as well as between base +145 and base +878 in relation to the transcription start site (TSS). +1: ST6GAL1 TSS. Orange and yellow boxes illustrating gene transcription-relevant regulatory elements statistically identified by using the Genomatix data base (http://www.genomatix.de/): BRE: Transcription factor II B (TFIIB) recognition element (score: 1.0, position in relation to TSS: 282–288 bp); SP4: Ubiquitous GC-Box factors SP1/GC recognition element for SP4 TF (score: 0.89, position in relation to TSS: 287–303 bp). (B) Representative MSP analysis illustrating the ST6GAL1 promoter methylation status of human bladder cell lines RT112, RT4, J82 and UROtsa. Band labels with U and M represent an unmethylated and methylated DNA locus. Bisulphite-converted unmethylated, genomic (U-co) and polymethylated, genomic (M-co) DNA were used as positive controls. NTC: non-template control. (C) Comparison of ST6GAL1 mRNA expression of human bladder cell lines showing an unmethylated ST6GAL1 promoter hypermethylation (UROtsa and RT4) with ST6GAL1 methylated J82 and RT112 cells. Vertical lines: + s.e.m. (D) DNA demethylation of the ST6GAL1 promoter correlates with ST6GAL1 re-expression in vitro. Real-time PCR of ST6GAL1 mRNA expression demonstrated a clear ST6GAL1 re-expression after treatment with both DAC (+) and TSA (+) only in the RT112 bladder cell line. Non-treated cells were set to 1. Error bars: + s.e.m.
MSP analysis showed indeed distinct DNA methylation in the human bladder cancer cell line J82 and RT112 at the ST6GAL1 promoter region. Of importance the ST6GAL1 promoter region of the immortalized bladder cell line UROtsa originally derived from normal urothelium remained unmethylated. Reflecting the variance seen in our tumor population, the RT4 papillary-invasive cell line showed no ST6GAL1 promoter methylation (Figure 3B). This unmethylated configuration of the ST6GAL1 promoter correlated with a strong ST6GAL1 expression in both cell lines UROtsa and RT4, whereas a weak expression in J82 and a nearly complete loss of ST6GAL1 mRNA expression in RT112 cancer cells was observed (Figure 3C).
In vitro demethylation assays of human bladder cell lines (RT112, RT4, J82 and UROtsa) were performed to further underscore the functional association between ST6GAL1 promoter methylation and gene transcription. Demethylation application led to an upregulation of ST6GAL1 mRNA expression approximately by 18,000 fold in highly methylated RT112 tumor cells (Figure 3D). ST6GAL1 mRNA expression in J82 cells was solely marginal re-expressed after DAC and TSA treatment. ST6GAL1 mRNA in both normal-like UROtsa and RT4 bladder tumor cells harboring an unmethylated ST6GAL1 promoter served as control and were not further inducible by DAC/TSA (Figure 3D). These data indicate that epigenetic configurations at the ST6GAL1 promoter region are involved in the regulation of ST6GAL1 expression.
ST6GAL1 promoter methylation is tightly associated with ST6GAL1expression loss in primary bladder tumors
Based on the promoter methylation in bladder cancer cell lines, the ST6GAL1 promoter methylation in primary human bladder cancer samples including CIS (n = 82) was analyzed using MSP technology. NU tissues (n = 23) served as control. All analyzed NU tissues showed unmethylated ST6GAL1 promoters; representative MSP results demonstrating an aberrant ST6GAL1 promoter region methylation status in bladder tumors are shown in Figure 4A. Overall, ST6GAL1 promoter methylation was detected in 32.9% (27/82) of bladder cancer samples. Upon closer examination with respect to the individual growth forms, the methylation frequency in invasive tumors was 53.6% (22/41), whereas only one out of 12 (8.3%) of the CIS samples showed aberrant ST6GAL1 promoter methylation. The non-invasive pTa phenotype displayed a methylation frequency of 13.8% (4/29). Such disparity between the two growth forms is not surprising, as findings by Wolff et al. suggested a general pattern of hypomethylation in noninvasive urothelial tumors [27].
ST6GAL1 promoter hypermethylation in primary human bladder cancer is associated with loss of ST6GAL1 mRNA expression. (A) Representative MSP results of the ST6GAL1 promoter methylation status in four papillary non-invasive pTa low grade tumors (PAP) as well as four invasive high grade bladder cancer (INV) samples in comparison to four normal tissue specimens (NU). Bands labeled with U and M show unmethylated and methylated DNA, respectively. Percent values reflect methylation frequency (M frequency). Bisulphite-converted unmethylated, genomic (U-co) and polymethylated, genomic (M-co) DNA were used as positive controls. DNA control: genomic, non- bisulphite-converted DNA, NTC: non-template control. (B) Box plot illustrating ST6GAL1 mRNA downregulation according to hypermethylated ST6GAL1 promoter status in bladder cancer (UC). (U): unmethylated tumors. (M): Methylated tumors. Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak and minimum; *p < 0.05.
Subsequently we correlated these methylation results with the ST6GAL1 mRNA expression in order to determine whether the promoter methylation was responsible for the loss of gene expression in muscle-invasive tumors. Unmethylated NU tissues served as a control (expression level set to 1). Compared to these, unmethylated UC tumors showed a median expression rate of 1.512. In contrast methylated bladder tumors showed a significant (p < 0.05) reduction in ST6GAL1 mRNA expression down to 0.338 (Figure 4B). The significance of the correlation between loss of ST6GAL1 gene expression and its promoter methylation was statistically confirmed by using a Fisher’s exact test (p = 0.022) (Table 4).
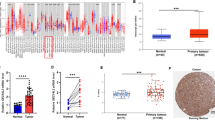
In order to strengthen our findings, we analyzed ST6GAL1 promoter methylation and gene expression in a dataset of independent studies (The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov)), in total representing 184 different bladder cancer samples (for cohort characteristics see Additional file 1: Table S1). Based on the TCGA data we verified downregulation of ST6GAL1 gene expression in bladder cancer in comparison to normal bladder tissues (Figure 5A). Furthermore, ST6GAL1 promoter hypermethylation of six CpG sites (located from -98 bp to +584 bp with respect to the TSS, i.e. covering the MSP analyzed region) was confirmed in bladder cancer samples underscoring a homogenous methylation pattern within the ST6GAL1 promoter locus (Figure 5B) compared to normal solid tissues. Importantly, a highly significant (p < 0.001) inverse correlation (Spearman coefficient: -0.733) of aberrant ST6GAL1 promoter methylation and ST6GAL1 mRNA expression was verified in this dataset (Figure 5C).
Tumor-specific ST6GAL1 promoter hypermethylation is associated with ST6GAL1 gene silencing in an independent TCGA data set. (A) ST6GAL1 expression in bladder tumor samples from the TCGA data portal. Red: high expression, black: mean expression and green: low expression. Left panel: sample type (dark grey: primary tumor; white: solid normal tissues). Right panel: ST6GAL1 mRNA expression. (B) DNA methylation of the ST6GAL1 promoter analyzed in bladder cancer samples from TCGA data portal. Red: high methylation, white: mean methylation; blue: low methylation. Right panel: sample type (dark grey: primary tumor; white: solid normal tissues). Left panels: values of ST6GAL1 DNA methylation for each available CG number. The relative positions of six analyzed CpG duplets within the ST6GAL1 promoter region covering the MSP analyzed region (+224 to +346) are indicated. +1: ST6GAL1 TSS. CG25372568 position in relation to the stated TSS: +272. (C) Inverse correlation of ST6GAL1 mRNA expression and its DNA methylation status in primary bladder cancer samples. ρ: Spearman correlation coefficient.
Discussion
Accumulating evidence shows that ST6GAL1 is aberrantly expressed in various cancer entities such as colon, breast, and epithelial tumor types [14, 17], and most studies propose an oncogenic role for ST6GAL1 [16–20, 28]. However, its role in tumorigenesis remains controversial [21]. Up to date, knowledge about a possible role of ST6GAL1 in human bladder cancer is still lacking. In the present study we aimed to describe for the first time ST6GAL1 expression and regulation in human bladder carcinogenesis.
To begin, we revealed a differential ST6GAL1 mRNA expression in bladder tumors compared to normal urothelium. Using real-time PCR, we were able to consistently show ST6GAL1 mRNA expression in normal urothelial tissue. Initial analysis of a mixed-stage tumor cohort failed to reveal a clear pattern of up- or downregulation in ST6GAL1 expression. However, when these samples were classified according to subtypes/stages, two distinct patterns emerged. Well-differentiated non-invasive papillary tumors were predominately characterized by an increase in ST6GAL1 expression, while invasive tumors (pT2-4) displayed a significant decrease in ST6GAL1 expression. Such disparity is, however, hardly surprising as both tumor forms are characterized by a unique molecular profile, which extends to the epigenetic level [8, 10, 29]. In addition, Seales et al. showed that expression of oncogenic RAS in HD3 colonocytes caused an increase in α-2,6-sialylation of β1-integrins by ST6GAL1, and the expression of dominant-negative RAS results in decreased sialylation [30]. Therefore, an upregulation of ST6GAL1 in papillary tumors, which often carry a mutated RAS gene [7] can be viewed in this context. Thereafter, using patient matched samples, a sequential loss of ST6GAL1 could be demonstrated throughout the course of bladder cancer development beginning in the CIS stage and continuing to invasive disease when compared with the corresponding normal urothelium. Consistent with these findings, we observed weak ST6GAL1 protein expression in invasive bladder tumors. Indeed, more than one fourth of high-grade cancer tissues showed complete loss of ST6GAL1 protein. These observations seem to contradict those by Swindall and colleagues, who reported a general upregulation of ST6GAL1 in epithelial tumors; however, their data was based on a very small sample population of ovarian, stomach and prostate cancers. Most notably, bladder cancer was not included in this cohort [28]. Of clinical importance, in our comprehensive sample collection, ST6GAL1 expression loss correlated with clinico-pathological metastasis, i.e. regional lymph node invasion, implying a possible tumor suppressive role for ST6GAL1 in advanced bladder cancer stages.
It is well known that epigenetic alterations, such as DNA methylation, are common mechanisms contributing to tumorigenesis and tumor progression [29, 31, 32]. Analysis of aberrant DNA methylation has rapidly garnered interest in cancer risk assessment, diagnosis, as well as prognosis and therapy monitoring [33, 34]. The progressive increase in de novo methylation of CpG islands in urothelial carcinoma suggests that epigenetic gene silencing is involved in the development of UC [10]. Interestingly, Fleischer et al. has previously shown that the ST6GAL1 promoter sequence contains distinct CpG islands whose DNA methylation caused loss of ST6GAL1 expression in breast cancer [25]. As such, we sought to determine whether promoter methylation could be responsible for ST6GAL1 gene inactivation in bladder cancer as well. Indeed, we identified a tumor-specific ST6GAL1 promoter hypermethylation, which appeared to be more frequently associated with poorly differentiated and advanced tumor stages. Importantly, a highly significant inverse correlation between ST6GAL1 expression and ST6GAL1 promoter hypermethylation was observed in these bladder tumors. It appears that loss of ST6GAL1 expression in bladder cancer is not only a common event, but also tightly associated with epigenetic changes within the ST6GAL1 promoter region. This hypothesis was strengthened using an independent bladder tumor data set based on The Cancer Genome Atlas (TCGA) portal. Finally, demethylating treatment of methylated ST6GAL1 bladder cancer cell lines with 5-aza-2-deoxycytidine clearly restored ST6GAL1 expression, thus functionally confirming a strong correlation between ST6GAL1 expression and its promoter hypermethylation. Our findings give the first indications that this major epigenetic mechanism could be important in the regulation of ST6GAL1 expression in bladder cancer as well. Bearing in mind that this is a known mechanism to mediate the silencing of tumor suppressor genes, a possible tumor suppressive role of ST6GAL1 in the course of bladder cancer progression could be hypothesized even though further studies are needed to analyze, in depth, ST6GAL1 function in human bladder cancer subtypes.
Conclusions
Our study characterizes for the first time ST6GAL1 expression and regulation during bladder cancer development. ST6GAL1 loss is caused by aberrant ST6GAL1 promoter methylation potentially indicating a tumor suppressive role in bladder carcinogenesis. Further investigation is warranted in order to more precisely assess the mechanism by which ST6GAL1 contributes to bladder cancer formation, progression and invasion.
Authors’ information
Pia Antony and Michael Rose are equal first authors.
Nadine T Gaisa and Edgar Dahl are equal co-senior authors.
Abbreviations
- ST6GAL1:
-
Beta-galactosamide alpha-2,6-sialyltransferase 1
- mRNA:
-
Messenger ribo nucleic acid
- n:
-
Number
- NU:
-
Normal urothelium
- pTa:
-
Papillary non-invasive tumors
- CIS:
-
Carcinoma in situ; pTis
- pT2-4:
-
Muscle invasive tumors
- PCR:
-
Polymerase chain reaction
- TCGA:
-
The Cancer Genome Atlas
- HG:
-
High grade
- LG:
-
Low grade
- IRS:
-
Immunoreactive score
- MSP:
-
Methylation specific PCR
- DNA:
-
Desoxyribonucleic acid
- UC:
-
Urothelial cell cancer
- FGFR3:
-
Fibroblast growth factor receptor 3
- RAS:
-
Rat sarcoma
- TP53:
-
Tumor protein 53
- RB:
-
Retinoblastoma
- CMP:
-
Cystidine monophosphate
- TNFR1:
-
Tumor necrosis factor receptor 1
- ATCC:
-
American Type Culture Collection
- USA:
-
United States of America
- RWTH:
-
Rheinisch Westfälisch Technische Hochschule
- EK:
-
Ethics committee
- cDNA:
-
Copy number desoxyribonucleic acid
- CA:
-
California
- WI:
-
Wisconsin
- GAPDH:
-
Glyeradehyde 3-phosphate dehydrogenase
- MSP:
-
Methylation specific PCR
- EDTA:
-
Tris ethylenediaminetetraacetic acid
- UV:
-
Ultraviolet
- dNTP:
-
Desoxyribonucleosidtriphosphate
- DAC:
-
5-aza-2′-deoxycytidine
- TSA:
-
Trichostatin A
- FFPE:
-
Formalin fixed paraffin embedded
- DAB:
-
3-3′ diaminobenzidine
- FC:
-
Fold change
- G1:
-
Well differentiated
- G2:
-
Moderately differentiated
- G3:
-
Poorly differentiated
- WHO:
-
World Health Organization
- s.e.m.:
-
Standard error of the margin
- PAP:
-
Papillary non-invasive tumors
- INV:
-
Invasive tumors
- U:
-
Unmethylated
- M:
-
Methylated preparation.
References
Parkin DM, Bray F, Ferlay J, Pisani P: Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001, 94: 153-156. 10.1002/ijc.1440.
Reuter VE: Bladder. Risk and prognostic factors-A pathologist’s perspective. Urol Clin North Am. 1999, 26: 481-492. 10.1016/S0094-0143(05)70196-8.
Castillo-Martin M, Domingo-Domenech J, Karni-Schmidt O, Matos T, Cordon-Cardo C: Molecular Pathways of urothelial development and bladder tumorigenesis. Urol Oncol. 2010, 28: 401-408. 10.1016/j.urolonc.2009.04.019.
McConkey DJ, Lee S, Choi W, Tran M, Majewski T, Lee S, Siefker-Radtke A, Dinney C, Czerniak B: Molecular genetics of bladder cancer: emerging mechanisms of tumor initiation and progression. Urol Oncol. 2010, 28: 429-440. 10.1016/j.urolonc.2010.04.008.
Goebbell PJ, Knowles MA: Bladder cancer or bladder cancers? Genetically distinct malignant conditions of the urothelium. Urol Oncol. 2010, 28: 409-428. 10.1016/j.urolonc.2010.04.003.
Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre-Garau X, Chopin D, Thiery JP, Radvanyi F: Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999, 23: 18-20.
Al Hussain TO, Akhtar M: Molecular basis of urinary bladder cancer. Adv Anat Pathol. 2013, 20: 53-60. 10.1097/PAP.0b013e31827bd0ec.
He F, Mo L, Zheng XY, Hu C, Lepor H, Lee EY, Sun TT, Wu XR: Deficiency of pRb family proteins and p53 in invasive urothelial tumorigenesis. Cancer Res. 2009, 69: 9413-9421. 10.1158/0008-5472.CAN-09-2158.
Cairns P, Proctor AJ, Knowles MA: Loss of heterozygosity at the RB locus is frequent and correlates with muscle invasion in bladder carcinoma. Oncogene. 1991, 6: 2305-2309.
Bilgrami SM, Qureshi SA, Pervez S, Abbas F: Promotor hypermethylation of tumor suppressor genes correlates with tumor grade and invasiveness in patients with urothelial bladder cancer. Springerplus. 2014, 3: 78-10.1186/2193-1801-3-78. doi:10.1186/2193-1801-3-178. eCollection 2014
Dall’Olio F, Malagolini N, Trinchera M, Chiricolo M: Sialosignaling: sialyltransferases as engines of self-fueling loops in cancer progression. Biochim Biophys Acta. 1840, 2014: 2752-2764.
Schultz MJ, Swindall AF, Bellis SL: Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metastasis Rev. 2012, 31: 501-518. 10.1007/s10555-012-9359-7.
Zhuo Y, Bellis SL: Emerging role of alpha2,6-sialic acid as a negative regulator of galectin binding and function. J Biol Chem. 2011, 286: 5935-5941. 10.1074/jbc.R110.191429.
Swindall AF, Bellis SL: Sialylation of the Fas death receptor by ST6Gal-I provides protection against Fas-mediated apoptosis in colon carcinoma cells. J Biol Chem. 2011, 286: 22982-22990. 10.1074/jbc.M110.211375.
Liu Z, Swindall AF, Kesterson RA, Schoeb TR, Bullard DC, Bellis SL: ST6Gal-I regulates macrophage apoptosis via alpha2-6 sialylation of the TNFR1 death receptor. J Biol Chem. 2011, 286: 39654-39662. 10.1074/jbc.M111.276063.
Zhu Y, Srivatana U, Ullah A, Gagneja H, Berenson CS, Lance P: Suppression of a sialyltransferase by antisense DNA reduces invasiveness of human colon cancer cells in vitro. Biochim Biophys Acta. 2001, 1536: 148-160. 10.1016/S0925-4439(01)00044-8.
Lin S, Kemmner W, Grigull S, Schlag PM: Cell surface alpha 2,6 sialylation affects adhesion of breast carcinoma cells. Exp Cell Res. 2002, 276: 101-110. 10.1006/excr.2002.5521.
Seales EC, Jurado GA, Brunson BA, Wakefield JK, Frost AR, Bellis SL: Hypersialylation of beta1 integrins, observed in colon adenocarcinoma, may contribute to cancer progression by up-regulating cell motility. Cancer Res. 2005, 65: 4645-4652. 10.1158/0008-5472.CAN-04-3117.
Shaikh FM, Seales EC, Clem WC, Hennessy KM, Zhuo Y, Bellis SL: Tumor cell migration and invasion are regulated by expression of variant integrin glycoforms. Exp Cell Res. 2008, 314: 2941-2950. 10.1016/j.yexcr.2008.07.021.
Bresalier RS, Rockwell RW, Dahiya R, Duh QY, Kim YS: Cell surface sialoprotein alterations in metastatic murine colon cancer cell lines selected in an animal model for colon cancer metastasis. Cancer Res. 1990, 50: 1299-1307.
Park JJ, Yi JY, Jin YB, Lee YJ, Lee JS, Lee YS, Ko YG, Lee M: Sialylation of epidermal growth factor receptor regulates receptor activity and chemosensitivity to gefitinib in colon cancer cells. Biochem Pharmacol. 2012, 83: 849-857. 10.1016/j.bcp.2012.01.007.
Wild PJ, Herr A, Wissmann C, Stoehr R, Rosenthal A, Zaak D, Simon R, Knuechel R, Pilarsky C, Hartmann A: Gene expression profiling of progressive papillary noninvasive carcinomas of the urinary bladder. Clin Cancer Res. 2005, 11: 4415-4429. 10.1158/1078-0432.CCR-05-0259.
Remmele W, Stegner HE: Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe. 1987, 8: 138-140.
Cancer Genome Atlas Research Network: Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014, 507: 315-322. 10.1038/nature12965.
Fleischer T, Edvardsen H, Solvang HK, Daviaud C, Naume B, Børresen-Dale AL, Kristensen VN, Tost J: Integrated analysis of high-resolution DNA methylation profiles, gene expression, germline genotypes and clinical endpoints in breast cancer patients. Int J Cancer. 2014, 134: 2615-2625. 10.1002/ijc.28606.
Li LC, Dahiya R: Methpimer: designing primers for methylation PCRs. Bioinformatics. 2002, 18: 1427-1431. 10.1093/bioinformatics/18.11.1427.
Wolff EM, Chihara Y, Pan F, Weisenberger DJ, Siegmund KD, Sugano K, Kawashima K, Laird PW, Jones PA, Liang G: Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res. 2010, 70: 8169-8178. 10.1158/0008-5472.CAN-10-1335.
Swindall AF, Londoño-Joshi AI, Schultz MJ, Fineberg N, Buchsbaum DJ, Bellis SL: ST6GAL1 protein expression is upregulated in human epithelial tumors and correlates with stem cell markers in normal tissues and colon cancer cell lines. Cancer Res. 2013, 73: 2368-2378. 10.1158/0008-5472.CAN-12-3424.
Sanchez-Cabayo M: Hypermethylation in bladder cancer. Tumor Biol. 2012, 33: 347-361. 10.1007/s13277-011-0310-2.
Seales EC, Jurado GA, Singhal A, Bellis SL: Ras oncogene directs expression of a differentially sialylated, functionally altered b1 integrin. Oncogene. 2003, 22: 7137-7145. 10.1038/sj.onc.1206834.
Ushijima T: Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005, 5: 223-231. 10.1038/nrc1571.
Kim WJ, Kim YJ: Epigenetic biomarkers in urothelial bladder cancer. Exp Rev Mol Diagn. 2009, 9: 259-269. 10.1586/erm.09.5.
Baylin SB, Jones PA: A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011, 11: 726-734. 10.1038/nrc3130.
Friedrich MG, Chandrasoma S, Siegmund KD, Weisenberger DJ, Cheng JC, Toma MI, Huland H, Jones PA, Liang G: Prognostic relevance of methylation markers in patients with non-muscle invasive bladder carcinoma. Eur J Cancer. 2005, 41: 2769-2778. 10.1016/j.ejca.2005.07.019.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/14/901/prepub
Acknowledgements
The authors are grateful for the support of the study by all staff of the Department of Urology University Hospital Aachen providing samples and data of patients, and the excellent technical assistance of Sevim Alkaya, Roswitha Davtalab and Sonja von Serényi. This project was funded by START-funds of the Medical Faculty of the RWTH Aachen University (NTG grant number: 149/08).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MR, RK, NTG and ED participated in the design of the study. PA and NTG carried out the experimental data acquisition. PA, MR and NTG performed data analyses. AH provided tumor samples and clinical data. PA and MR wrote the manuscript. All authors read, critically revised, and approved the final manuscript.
Electronic supplementary material
12885_2014_5068_MOESM1_ESM.doc
Additional file 1: Table S1: Clinico-pathological parameters of 184 bladder cancer specimens (TCGA) analyzed in this study. (DOC 43 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Antony, P., Rose, M., Heidenreich, A. et al. Epigenetic inactivation of ST6GAL1 in human bladder cancer. BMC Cancer 14, 901 (2014). https://doi.org/10.1186/1471-2407-14-901
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2407-14-901