Abstract
Background
DNA fingerprinting by IS6110-RFLP has shown a high incidence of Mycobacterium tuberculosis isolates having no and low copies of the insertion sequence in Kerala, South India. Amplified Fragment Length Polymorphism (AFLP) would scan the entire genome rather than a few repetitive elements, we thought that this technique would help us in differentiating the large reservoir of isolates from an endemic region. Here we evaluate the ability of Amplified Fragment Length Polymorphism (AFLP) to type clinical isolates.
Methods
Fifty clinical isolates of M. tuberculosis were analysed by conventional radioactive AFLP and IS6110- RFLP. M. bovis, M. bovis BCG and two non tuberculous mycobacteria were also analysed to see species specific differences generated by AFLP. Cluster analysis was performed using the AFLP profile that showed the maximum polymorphism within M. tuberculosis and this was compared to the number of copies of IS6110 insertions.
Results
For AFLP, out of ten primer pairs tested, the EO/MC pair generated maximum polymorphism among the clinical isolates of M. tuberculosis. The similarity between the isolates ranged between 88 and 99.5%. Majority (nearly 85%) of the 'low copy' IS6110 isolates clustered together, while the rest clustered irrespective of the copy numbers. AFLP could show rare differences between isolates of M. tuberculosis, M. bovis and M. bovis BCG. The AFLP profiles for non-tuberculous mycobacteria were highly different from those of M. tuberculosis.
Conclusion
Polymorphism generated by AFLP within the M. tuberculosis species is limited and hence AFLP alone seems to have limited use in fingerprinting the isolates in Kerala. The combined use of AFLP and IS6110-RFLP showed relatively better differentiation of 'high copy' IS6110 isolates, but failed to differentiate the 'low copy' isolates. However, the technique may be efficient in inter-species differentiation, and hence potentially useful in identifying and developing species- specific markers.
Similar content being viewed by others
Background
Tuberculosis claims about 500, 000 deaths annually in India and the disease is endemic in Kerala, a small state in the southwest [1]. DNA fingerprinting by RFLP-hybridization based on the insertion sequence IS6110 [2] showed a high prevalence of 'no copy' and single copy isolates of Mycobacterium tuberculosis in Kerala [3]. This, and other studies point out that IS6110-based methods have limited use in regions where there is a high incidence of strains carrying few copies of IS6110 [4, 5]. Spoligotyping, the next widely used fingerprinting method for M. tuberculosis complex organisms can efficiently differentiate 'low copy' (5 or fewer copies of IS6110) strains but again is not as efficient as RFLP for 'high copy' (6 or more copies of IS6110) strains [6, 7]. Fingerprinting techniques such as Random Amplification of Polymorphic DNA (RAPD) and Amplified Fragment Length Polymorphism (AFLP) would scan the entire genome rather than a few repetitive elements, and since AFLP can be applied to any genome irrespective of its complexity [8], we thought that this technique would help us in differentiating the large reservoir of isolates from an endemic region. AFLP has been used for strain typing within different bacterial genera [9] and has also been evaluated in inter- and intra-species typing of mycobacteria [10–13]. In the present study, we analyzed clinical isolates of M. tuberculosis from Kerala using AFLP and looked at the ability of the technique to further differentiate isolates typed by IS6110-RFLP. The ability of AFLP to provide inter-species differentiation was used to prove the validity of the methodology adopted by us.
Methods
Clinical isolates and laboratory strains
Fifty clinical isolates of Mycobacterium tuberculosis, isolated from TB patients and identified based on their growth characteristics, colony morphology and biochemical tests [14] were used in this study. The Laboratory strains of mycobacteria were M. tuberculosis H37Rv, H37Ra, M. bovis and M.bovis BCG. Two atypical mycobacteria obtained from clinical samples were also included in the study. Using INNO-LiPA Mycobacteria (LiPA; Innogenetics, Zwijnaarde, Belgium), one was identified as M. chelonae, while the other could be identified to genus level only.
The isolates used in this investigation were collected by our laboratory as a part of a study to screen for anti-TB drug sensitivity and became part of our local repository. Since this was a retrospective study on the isolates already available with us, it was not put up for any clearance to an ethical committee.
Genomic DNA
Genomic DNA was isolated from mycobacteria using CTAB method as reported earlier [2].
AFLP analysis
AFLP adapters and primers [8] were custom synthesized by Sigma Genosys (Table 1). AFLP analysis was carried out as described by Vos et al [8]. Briefly, about 200 ng genomic DNA from each strain/isolate was digested with EcoR I and Mse I (New England Biolabs, Beverly, MA). Each sample of digested DNA was ligated to EcoR I and Mse I adapters (5 pmol and 50 pmol respectively). The ligated mixture was diluted to 1:10 ratio in 1X TE buffer and used as template for PCR. Just before the PCR, EcoR I primer was labeled with γ-32P (3000 Ci/mmol ATP), using T4 polynucleotide kinase (NEB). Five μl template DNA, 30 ng Mse I primer, 5 ng labeled EcoR I primer, were used for each 20 μl PCR reaction. PCR was carried out in an iCycler (Bio-Rad Labs, Hercules, CA). As described by Vos et al (8), the first cycle was carried out at 94°C for 30 sec, 65°C for 30 sec and 72°C for 60 sec. The annealing temperature was lowered each cycle by 0.7°C during the next 12 cycles (touch down phase of 13 cycles) and finally 23 cycles were carried out at 94°C for 30 sec, 56°C for 30 sec and 72°C for 60 sec.
The PCR products were analyzed by urea-PAGE. Gel images were visualized in a Phosphor imager (Personal Molecular Imager FX, Bio-Rad, Hercules, CA).
IS6110-RFLP
This was performed according to the standard protocol [2].
Cluster analysis was performed using similarity coefficient based on bands that showed differences among isolates. Bionumerics software v 3 (Applied Maths, Belgium) was used to create UPGMA dendrogram.
Results
AFLP analysis
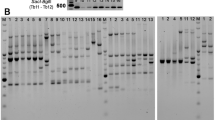
The initial AFLP analysis of twenty five clinical isolates of M. tuberculosis using ten primer combinations showed that, combinations having a single selective primer yielded 40–50 bands, while any two selective primers, when combined, yielded only 15–35 bands. A band, if either absent or present in 10–90% of the isolates tested was considered to be polymorphic for the clinical isolates. Accordingly, there was relatively low polymorphism within the species (an average 4.27 % of the total number of bands/primer pair was polymorphic). But for two isolates of non-tuberculous mycobacteria, the AFLP profiles for all the primer combinations were highly different from those of M. tuberculosis (Figure 1). It was observed that the use of a selective primer pair increased the number of polymorphic bands while reducing the total number of bands obtained, whereas nonselective primer pairs had the opposite effect. Therefore, we analysed fifty clinical isolates of M. tuberculosis along with the laboratory strains of M. tuberculosis, M. bovis and BCG using four primer pairs, with each pair having one selective and one non-selective primer (EO/MT, EO/MG, EO/MA and EO/MC). Among these primer combinations, EO/MC combination showed maximum polymorphism within the M. tuberculosis species. The AFLP profiles for isolates of M. bovis and BCG did not vary much from those of M. tuberculosis, although there were a few bands that may be species-specific. In addition, there were a few differences between M. bovis and the BCG vaccine strain. Figure 2 shows the AFLP profiles for two primer combinations (EO/MC and EO/MT) that showed strain-specific differences.
AFLP profiles generated by primer combinations EO/MT (panel A) and EG/MC (panel B). Lanes1-8: clinical isolates of M. tuberculosis, lanes 9&10: non tuberculous mycobacteria. The non-tuberculous mycobacteria have a very different profile from each other as well as from M. tuberculosis isolates. The primer pair EG/MC (panel B) shows fewer bands and more differences between the isolates as compared to EO/MT primer pair (panel A)
Comparison of AFLP with IS6110- RFLP
Since we obtained maximum differences using the EO/MC primer pair, we used the AFLP profile obtained with this primer pair to construct a dendrogram (Figure 3) and compared this with the IS6110 fingerprinting data. Seventeen out of fifty isolates tested had IS6110 copy numbers ranging from 6 to 17. The remaining 33 were 'low copy' (0–5 copies) isolates, out of which 21 isolates had one, and three had no copy at all. This was in line with our earlier report on the high incidence of 'low copy isolates' in Kerala [3]. Cluster analysis showed three major clusters with the lowest similarity between them being 88%. The largest cluster (Cluster1) contained M. tuberculosis H37Rv, H37Ra and most of the clinical isolates. The subcluster 1a contained H37Rv, H37Ra and 7 clinical isolates, of which two were 'no copy' and five had 9 or 10 copies. Similarity within this subcluster ranged from 96–99.3%. The subcluster 1b had 28 (85%) 'low copy' and four high copy isolates, with similarity ranging from 96.4 to 99.5%. Cluster 2 contained only M bovis and BCG which are known to be similar. Eight 'high copy' isolates (with copies varying from 7 to 17) along with two single copy isolates formed Cluster 3. This cluster showed least intra-cluster similarity with values ranging from 94.2–98.5%. Overall, clusters 1 and 2 had 92.8% similarity to each other with only 88% similarity to cluster 3. The other 3 primer pairs showed fewer polymorphic bands, showed very little differentiation among the different isolates and were not considered for numerical analysis (data not shown).
Discussion
In the present study we have used conventional radioactive AFLP in order to evaluate its potential to differentiate the M. tuberculosis isolates of Kerala. This is an endemic region where we have reported a high incidence of M. tuberculosis strains with low numbers of IS6110 which makes it difficult to be type them [3]. We used this method instead of multiplex Fluorescent AFLP, as we were also interested in developing AFLP-derived markers for the characterization of the Kerala isolates. However, the data presented in this paper is restricted to AFLP profiles and their comparison to IS6110 copy number data for a set of 50 isolates, of which 33 were 'low copy' isolates. Initially, our results on twenty-five clinical isolates showed 0–3 polymorphic bands per primer pair while using ten primer combinations. Using EcoR I/Mse I enzyme combination, approximately 4% of the total number of bands/primer combination was polymorphic for M. tuberculosis. There have been earlier reports on AFLP analysis of Mycobacterium tuberculosis. A study using multiplex FAFLP employing EcoR I and Mse I enzymes and four primer combinations had shown that 28% of the total number of fragments obtained were polymorphic [11]. In contrast, another study using radioactive AFLP and a different enzyme combination (Apa I/Taq I), demonstrated poor discrimination within the M. tuberculosis complex [10]. The discrepancy between these studies could be due to the choice of restriction enzymes and the methodology. FAFLP involves multiplexing which is not feasible with the radioactive methodology. Therefore, we have used a combination of the above methodologies using EcoR I/Mse I enzyme combination and conventional radioactive AFLP. Even though one may screen other enzyme combinations to increase polymorphism, our results suggest that radioactive AFLP is not an easy method for strain differentiation in endemic regions. However, AFLP could clearly differentiate M. tuberculosis from isolates of non-tuberculous mycobacteria. In our study, even though the close relationship between members of M. tuberculosis complex is reflected in AFLP analysis, the rare differences could also be detected. We characterized a few bands that differentiated M. tuberculosis and M. bovis/BCG by sequencing and identified three fragments belonging to RD1, RD4 and RD5 (results not shown). It has been reported that RD1 is absent only in BCG, while RD4-RD10 are deleted in both M. bovis and BCG [15]. Our results therefore correlate well with this data.
Comparing AFLP profile with IS6110 typing data showed that in AFLP analysis isolates clustered irrespective of the IS6110 copy number. The IS6110 'low copy' isolates were not well differentiated as nearly 85% of the low copy isolates clustered together (Fig 3, Cluster 1b). Therefore, it could be speculated that the low copy strains of Kerala are not very diverse. But all strains with a single copy of IS may not be identical, as they fell into different clusters (both 1 and 3) in the AFLP analysis. The high copy isolates were relatively better separated than the low copy isolates.
Conclusion
We conclude that conventional radioactive AFLP alone does not seem to be an efficient fingerprinting method for M. tuberculosis. The combined use of IS6110-RFLP and AFLP again can differentiate the 'high copy' isolates to some extent, but does not differentiate the 'low copy' isolates efficiently. However the technique provides inter-species differentiation within and outside the M. tuberculosis complex and therefore is useful in the development of species-specific markers. The latest addition to the fingerprinting methods for M. tuberculosis, MIRU-VNTR [16, 17], is efficient and is being adopted by several labs and we are currently analyzing our isolates using MIRU fingerprinting. But different fingerprinting techniques cluster the strains differently, and therefore generally appear to work independently of each other. Therefore, a unified fingerprinting system that can neatly pigeonhole all different strains is yet to be developed.
References
Radhakrishna G, Sumathi G: Performance of RNTCP in Himachal Pradesh and Kerala-a perspective comparison. NTI Bulletin. 2003, 39: 19-23. [http://medind.nic.in/nac/t03/i3/nact03i3c.shtml]
van Embden J, Cave M, Crawford J, Dale J, Eisenach K, Gicquel B, et al: Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardised methodology. J Clin Microbiol. 1993, 31: 406-409.
Radhakrishnan I, Manju YK, Ajay Kumar R, Sathish M: Implication of low frequency of IS6110 in fingerprinting field isolates of Mycobacterium tuberculosis from Kerala, India. J Clin Microbiol. 2001, 39: 1683-10.1128/JCM.39.4.1683.2001.
Yuen L, Ross B, Jackson K, Dwyer B: Characterization of Mycobacterium strains from Vietnamese patients by southern blot hybridization. J Clin Microbiol. 1993, 31: 1615-1618.
Das S, Paramasivan CN, Lowrie DB, Prabhakar R, Narayanan PR: IS6110 RFLP typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, South India. Tubercle Lung Dis. 1995, 76: 550-554. 10.1016/0962-8479(95)90533-2.
Goyal M, Saunders NA, van Embden JDA, Young DB, Shaw RJ: Differentiation of Mycobacterium tuberculosis isolates by spoligotyping and IS6110 restriction fragment polymorphism. J Clin Microbiol. 1997, 35: 647-651.
de la Salmoniere YG, Li HM, Torrea G, Bunschoten A, van Embden J, Gicquel B: Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J Clin Microbiol. 1997, 35: 2210-2214.
Vos P, Hogers R, Bleeker M, Reijans M, de Lee TV, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M: AFLP a new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23: 4407-4414. 10.1093/nar/23.21.4407.
Savelkoul PH, Aarts HJ, de Haas J, Dijkshoorn L, Duim B, Otsen M, Rademaker JL, Schouls L, Lenstra JA: Amplified-fragment length polymorphism analysis: the state of an art. J Clin Microbiol. 1999, 37: 3083-3091.
Huys G, Rigouts L, Chemlal K, Portaels F, Swings J: Evaluation of amplified fragment length polymorphism analysis for inter- and intraspecific differentiation of Mycobacterium bovis, M. tuberculosis, and M. ulcerans. J Clin Microbiol. 2000, 38: 3675-3680.
Goulding JN, Stanley J, Saunders N, Arnold C: Genome-sequence-based fluorescent amplified-fragment length polymorphism analysis of Mycobacterium tuberculosis. J Clin Microbiol. 2000, 38: 1121-1126.
van den Braak N, Simons G, Gorkink R, Reijans M, Eadie K, Kremers K, van Soolingen D, Savelkoul P, Verbrugh H, van Belkum A: A new high-throughput AFLP approach for identification of new genetic polymorphism in the genome of the clonal microorganism Mycobacterium tuberculosis. J Microbiol Meth. 2004, 56: 49-62. 10.1016/j.mimet.2003.09.018.
O'Shea B, Khare S, Bliss K, Klein P, Ficht TA, Adams LG, Rice-Ficht AC: Amplified Fragment Length Polymorphism Reveals Genomic Variability among Mycobacterium avium subsp. paratuberculosis isolates. J Clin Microbiol. 2004, 42: 3600-3606. 10.1128/JCM.42.8.3600-3606.2004.
Kubica GP: Identification of mycobacteria. Am Rev Respir Dis. 1973, 107: 9-21.
Gordon SV, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole ST: Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol Microbiol. 1999, 32: 643-655. 10.1046/j.1365-2958.1999.01383.x.
Supply P, Magdalena J, Himpens S, Locht C: Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol Microbiol. 1997, 26: 991-1003. 10.1046/j.1365-2958.1997.6361999.x.
Supply PE, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C: Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000, 36: 762-771. 10.1046/j.1365-2958.2000.01905.x.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2334/7/86/prepub
Acknowledgements
BVJ and ML are recipients of Research Fellowships from Council for Scientific and Industrial Research (CSIR), Government of India. This study received financial assistance under Program Support from the Department of Biotechnology, Government of India. The strains used in this study were collected under an SRF award of the Kerala State Council for Science, Technology and Environment to SM and AK. The type strains were kind gifts of Dr V M Katoch, JALMA, Agra and Dr S Vijaya, Indian Institute of Science, Bangalore and Tuberculosis Research Centre, Chennai.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
MK carried out the AFLP analysis, cluster analysis and drafted the manuscrift
IR and BVJ carried out the IS6110-RFLP analysis
ML has helped in genomic DNA isolation and PCR
AK helped in drafting the manuscript and did the critical review
SM conceived of the study, and participated in its design and coordination and helped to draft the manuscript
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Krishnan, M.Y., Radhakrishnan, I., Joseph, B.V. et al. Combined use of Amplified Fragment Length Polymorphism and IS6110 -RFLP in fingerprinting clinical isolates of Mycobacterium tuberculosis from Kerala, South India. BMC Infect Dis 7, 86 (2007). https://doi.org/10.1186/1471-2334-7-86
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2334-7-86