Abstract
Background
The yeast Xanthophyllomyces dendrorhous synthesizes astaxanthin, a carotenoid with high commercial interest. The proposed biosynthetic route in this organism is isopentenyl-pyrophosphate (IPP) → geranyleranyl pyrophosphate (GGPP) → phytoene → lycopene → β-carotene → astaxanthin. Recently, it has been published that the conversion of β-carotene into astaxanthin requires only one enzyme, astaxanthin synthase or CrtS, encoded by crtS gene. This enzyme belongs to the cytochrome P450 protein family.
Results
In this work, a crtR gene was isolated from X. dendrorhous yeast, which encodes a cytochrome P450 reductase (CPR) that provides CrtS with the necessary electrons for substrate oxygenation. We determined the structural organization of the crtR gene and its location in the yeast electrophoretic karyotype. Two transformants, CBSTr and T13, were obtained by deleting the crtR gene and inserting a hygromycin B resistance cassette. The carotenoid composition of the transformants was altered in relation to the wild type strain. CBSTr forms yellow colonies because it is unable to produce astaxanthin, hence accumulating β-carotene. T13 forms pale colonies because its astaxanthin content is reduced and its β-carotene content is increased.
Conclusion
In addition to the crtS gene, X. dendrorhous requires a novel gene, crtR, for the conversion of β-carotene to astaxanthin.
Similar content being viewed by others
Background
Carotenoids are natural pigments of yellow, orange or red color. More than 600 different chemical structures have been described to date [1]. They are terpenoids with the isopentenyl- pyrophosphate (IPP) molecule as the basic unit. Astaxanthin is a carotenoid with a high commercial interest due to its use as a food additive for trout and salmon flesh pigmentation in aquaculture [2]. Its biosynthesis is limited to a few microorganisms such as the microalgae Haematococcus pluvialis and the basidiomycete yeast Xanthophyllomyces dendrorhous [3, 4].
The first step in the biosynthesis of astaxanthin in X. dendrorhous is the isomerization of IPP into dimethylallyl pyrophosphate (DMAPP) by IPP isomerase, which is encoded by the idi gene [5]. Three IPP molecules are sequentially added to one DMAPP molecule, where the GGPP synthase (crtE gene) is involved, forming geranylgeranyl pyrophosphate (GGPP) [6]. Then, the condensation of two GGPP molecules produces phytoene, which corresponds to the first carotenoid synthesized in the pathway. This step is catalyzed by a bi-functional enzyme called phytoene-β-carotene synthase (PBS), which also has lycopene cyclase activity and is encoded by the crtYB gene [7]. Subsequently, phytoene goes through four successive desaturation reactions, catalyzed by the phytoene desaturase enzyme (crtI gene), producing lycopene [8]. Then, both lycopene ends are cyclized by the lycopene cyclase activity of the bi-functional PBS enzyme, forming β-carotene.
In the last step for the formation of astaxanthin from β-carotene, two enzymatic activities are required, ketolase, to incorporate a keto group in positions 4 and 4' of β-carotene, and hydroxylase, to introduce a hydroxyl group at positions 3 and 3' of β-carotene [9, 10]. Unlike other organisms, where two independent genes are needed for this step, in X. dendrorhous a single crtS gene has been isolated that encodes an astaxanthin synthase which can probably perform both activities [11–13]. However, it has been suggested that CrtS may require an auxiliary enzyme for the formation of astaxanthin from β-carotene [14]. The deduced CrtS amino acid sequence strongly suggests that the X. dendrorhous enzyme belongs to the cytochrome P450 protein family [12, 14].
Cytochrome P450 enzymes require an electron donor for the addition of oxygen-bearing functional groups to a substrate. In eukaryotes, this electron donor is cytochrome P450 reductase (CPR), encoded by the cpr gene [15]. Although several genes for different cytochrome P450 enzymes can exist in an organism, in most species only one cpr gene exists. Some exceptions have been observed in plants and zygomycetes that contain two or three cpr genes [16].
In another study, an Escherichia coli strain was transformed with three compatible plasmids. The first plasmid contained the necessary genes for the synthesis of β-carotene, the second contained the X. dendrorhous crtS gene, and the third contained the Saccharomyces cerevisiae cpr gene [12]. Through this system, it was possible to obtain oxygenated β-carotene derivatives, but not astaxanthin, suggesting the need for an appropriate CPR to complement the astaxanthin biosynthetic pathway. Prior to this study, a X. dendrorhous gene encoding for a CPR for astaxanthin biosynthesis had not been isolated. This paper describes for the first time the sequence of the X. dendrorhous crtR gene and its encoded polypeptide. Moreover, our results reveal that the crtR gene product is essential for astaxanthin biosynthesis in X. dendrorhous.
Methods
Strains, plasmids, media and enzymes
The strains and plasmids used in this study are listed in Tables 1 and 2. The wild type X. dendrorhous UCD 67–385 strain was used for genomic and cDNA library construction. E. coli DH-5α strain was used as a host for plasmid propagation and genomic libraries. Two genomic libraries were used in this study. The first library consisted of genomic DNA partially digested with BamHI and cloned into the BamHI site of the YIp5 vector [6]. The second library consisted of approximately 5 to 6 kb genomic DNA fragments digested with SalI and cloned into the SalI site of the pBluescript SK- vector. The cDNA library was constructed with the pBluescript II XR cDNA library construction kit from Stratagene [6].
crtR gene deletion bearing plasmids (pBsiWIΔcrtR::hph and pNdeIΔcrtR::hph) were built from pCPR1.3 (Table 2), which has the X. dendrorhous crtR gene. The deletions were created by digesting pCPR1.3 with BsiWI or NdeI and then filling the ends with Klenow polymerase to enable the ligation of the blunt ended hygromycin B resistance cassette (hph) [6] for the selection of X. dendrorhous transformants.
X. dendrorhous strains were grown at 22°C with constant agitation in YM medium (1% glucose, 0.3% yeast extract, 0.3% malt extract and 0.5% peptone) or MMV + 2% glucose medium [17]. UCD 67–385 strain fermentation was performed in a 12 l jar fermentor (New Brunswick) containing 8.8 l of MMV + 2% glucose medium, shaking at 300 rpm and 13 l/min sterile air injection. Antifoam agent (1520 US, Dow Corning) was automatically added when required. E. coli strains were grown with constant agitation at 37°C in Luria-Bertani (LB) medium and supplemented with 100 μg/ml ampicillin for plasmid selection and 40 μl of a 2% solution of X-gal (5-bromo-4chloro-3-indolyl-β-D-galactopyranoside) for recombinant clone selection [18]. For E. coli complementation experiments, the LB growth medium was supplemented with 10 μM hemine and 0.5 mM δ-aminolevulinic acid, which supports heme prosthetic group formation in E. coli [12, 19]. The expression was induced with 1 mM IPTG. Enzymes were purchased from Promega (TaqDNA pol, restriction enzymes, Klenow polimerase, M-MLV reverse transcriptase).
DNA amplification
Primers were purchased from Alpha DNA (Montreal, Canada) and are listed in Table 3. PCR reactions were performed in a final volume of 25 μl containing 2 U of TaqDNA pol, 2.5 μl of 10× Taq buffer, 0.5 μl of 10 mM dNTPs, 1 μl of 50 mM MgCl2, 1 μl of 25 μM of each primer and 10–20 ng DNA. Amplification was done in a DNA thermal cycler 2400 (Perkin-Elmer) as follows: initial denaturation at 95°C for 3 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, synthesis at 72°C for 3 min and a final extension step at 72°C for 10 min. Samples were kept at 4°C until checked. The amplicons were separated by 0.8% agarose gel electrophoresis in TAE buffer containing 0.5 μg/ml ethidium bromide [18] followed by DNA purification using glassmilk [20] for sequencing.
DNA sequence and bioinformatic tools
Nucleotide sequences were obtained from an ABI 3100 Avant genetic analyzer, using a DYEnamic ET terminator Kit (Amersham Bioscience). DNA sequences were analyzed with Vector NTI Suite 10 (Informax), CLUSTAL W 1.8 and BLAST programs. Protein analysis was performed with the online programs InterProScan http://www.ebi.ac.uk/InterProScan/ and TMPRED http://www.ch.embnet.org/software/TMPRED_form.html. Phylogenetic analysis was carried out using BioEdit 7.0.0 and Treecon 1.3b.
Pulse field gel electrophoresis
Chromosomal DNA was separated by contour-clamped homogeneous electric fields (CHEF) in a CHEF II BioRad system in 0.9% agarose gels in TBE 0.5× buffer (45 mM Tris-borate, 1 mM EDTA pH 8.0) at 14°C [21]. The pulses used were 90 s for 24 h, followed by 120 s for 24 h at 6 V/cm.
Southern blot hybridization
Southern blot hybridization was performed according to Sambrook and Russell [18]. crtR probe was obtained by PCR amplification with primers CPR3 and CPR7 of the crtR gene. hph probe was acquired from PCR amplification of the hph cassette using primers HF and HR. Probes were labeled with [32P]α-dCTP using the Promega prime-a-gene labeling system.
RNA extraction and cDNA synthesis
Total RNA extraction was performed according to a modified protocol of Chomczynski and Sacchi [22, 23]. The determination of the relative levels of crtS and crtR mRNAs was performed using a semi-quantitative RT-PCR method. The intensities of the crtS and crtR amplification bands were normalized with the intensity of the actin [24] amplification product [23, 25]. The amplicons were quantified with ImageJ 1.40 using a 100 bp DNA ladder (Fermentas) as the standard.
X. dendrorhoustransformation
X. dendrorhous transformation was performed by electroporation according to [26] and [27]. Electrocompetent cells were prepared from an exponential culture with DO600 nm = 1.2 grown in YM medium [6] and electroporated using a BioRad gene pulser × cell with PC and CE modules under the following conditions: 125 mF, 600 Ω, 0.45 kV. Transformant selection was performed in YM 1.5% agar plates supplemented with 10 μg/ml hygromycin B. The transformants were identified as X. dendrorhous by ITS1, 5.8 rRNA gene and ITS2 DNA sequences [28].
Pigment extraction and RP-HPLC
Carotenoid extraction was carried out from cellular pellets according to the acetone extraction method [29]. Carotenoids were quantified by absorbance at 465 nm using an absorption coefficient of A1% = 2,100. The analyses were performed in triplicate, and pigments were normalized relative to the dry weight of the yeast. Carotenoids were separated by RP-HPLC using a reverse phase RP-18 Lichrocart 125-4 (Merck) column with acetonitrile: methanol: isopropyl (85:10:5 v/v) as the mobile phase with a 1 ml/min flux under isocratic conditions. The elusion spectra were recuperated using a diode array detector.
Results and discussion
Cloning of crtRgene
Three cytochrome P450 reductase genes were used for primer design: from the zygomycete Cunninghamella echinulata [GenBank:AF195660], the ascomycete Saccharomyces cerevisiae [GenBank:D13788] and the basidiomycete Rhodotorula minuta [GenBank:AB055119]. Based on the conserved DNA segments, degenerate primers were designed (Table 3). PCR reactions were performed using genomic DNA from UCD 67–385 X. dendrorhous as a template and different combinations of forward and reverse primers. An amplicon of 1.6 kb was obtained when PCR reactions were performed with primers CPR3 and CPR7. This amplicon was completely sequenced and was found to be homologous to CPR genes by BLAST analysis. We named this gene crtR and designed specific crtR primers.
The YIp5 genomic library was screened for the X. dendrorhous crtR gene by PCR [6], and one recombinant plasmid carrying the crtR gene (pPR52.P.44) was isolated. This plasmid contained a DNA insert of approximately 12.8 kb, carrying six BamHI fragments of 5.4 kb, 4.4 kb, 0.9 kb (doublet), 0.8 kb and 0.4 kb. Each fragment was sub-cloned into the pBluescript SK- vector for sequencing. The sequence analysis showed that the crtR gene was in one end of the 5.4 kb fragment (pCPR1.3), but it was incomplete. The promoter region and part of the first exon were missing due to the presence of a BamHI site in the crtR gene.
A cDNA library was screened for the X. dendrorhous crtR cDNA, and a recombinant plasmid, pcXDA.44.29, was isolated, which contained a 2,680 bp insert corresponding to the cDNA of the crtR gene [GenBank:EU884134]. We used this cDNA sequence for specific primer design (primers CPREx1fwdb and CPREx1rev, Table 3) from the remaining crtR gene sequence, and this allowed the isolation of the pPR32.AA.51 plasmid from the YIp5 genomic library. pPR32.AA.51 contains the promoter and 5' region of the first exon of the crtR gene. Sequence analysis indicated that the 5' region of the crtR gene is at one end of the 4.5 kb BamHI DNA fragment.
The in silico DNA sequence analysis of the inserts from pPR32.AA.51 and pPR52.P.44 indicated that the crtR gene is entirely contained in a 5.9 kb SalI DNA fragment. This result was corroborated by Southern blot hybridization (data not shown). The recombinant plasmid pCPRB.6, which contains an insert of 5,896 bp that harbors the crtR gene, was isolated from a partial SalI genomic library. The complete crtR gene sequence [GenBank:EU884133] is displayed in Figure 1.
Graphical representation and sequence of the crtR gene from X. dendrorhous. The three crtR exons are highlighted in red and white letters. The restriction enzymes used in this work and their recognition sites are in parentheses. Underlined and bold black letters represent the donor and acceptor splice sites. The sequence is numbered in accordance with its graphical representation. The complete sequence of the DNA fragment that contains the crtR gene is in GenBank [GenBank:EU884133]. pCPR1.3 corresponds to the vector pBluescript SK- bearing the DNA fragment from site BamHI 4529 to BamHI 9895. crtR deletions were constructed from pCPR1.3.
Localization of the crtRgene in the yeast electrophoretic karyotype
To study the crtR gene's genomic organization, its localization in the electrophoretic karyotype of the UCD 67–385 strain was performed by Southern blot hybridization. The results showed that the crtR gene is in the first two chromosomal bands (Figure 2), where each band corresponds to a triplet [21]. As it was recently demonstrated that this strain is diploid [30], it is possible to conclude that the crtR gene is in at least two chromosomes and could be homologous polymorphic. In different X. dendrorhous strains, a high chromosomal polymorphism has been revealed [31], so it is possible that these homologous chromosomes have experienced genetic rearrangement, thus explaining their different sizes.
Sequence analysis of crtRgene
The crtR gene consists of three exons of 222, 317 and 1702 bp and two introns of 240 and 138 bp. This gene encodes a 746 amino acid protein (Figure 3) with a predicted molecular weight of 81.9 kDa. CrtR has 22 hydrophobic amino acids in its amino terminal region, a typical transmembrane segment of known CPRs that anchors the protein to the endoplasmic reticulum membrane in the cell [32]. CrtR also has recognizable CPR conserved domains such as a FMN binding domain, a FAD binding domain between amino acids 275 and 512, and a NAD(P)H binding domain between amino acids 582 and 709. The flavodoxin signature, which is only found in certain bacteria and algae, was identified between amino acids 80 and 222. However, the FMN binding domain of CPRs is homologous to bacterial flavodoxins [33]. Two putative P450-binding regions (P450-1 and P450-2) have been suggested [32], and one (P450-2) was studied by site-directed mutagenesis in rat CPR where the importance of acidic amino acid residues in this region was demonstrated [34]. The corresponding regions in CrtR were characterized by the presence of acidic amino acids (aspartic acid at positions: 112, 117, 122, 127, 210, 211, 218; glutamic acid at positions: 114, 115, 119, 126, 216, 217). The aspartic acid residues at position 210, 211 and 218 are extremely conserved in all known CPRs [32]. The P450 binding regions are located in the FMN binding domain. This is consistent with the electron flow from NAD(P)H to FAD to FMN and finally to the cytochrome P450 monooxygenase enzyme.
Deduced amino acid sequence from X. dendrorhous CrtR. (A) Graphical representation of CPR conserved domains in X. dendrorhous CrtR. (B) Deduced CrtR protein sequence. Conserved domains are shown in white text. TM: in italics and lower case. Highlighted in yellow: FMN BD, green: FAD BD and dark green: NAD(P)H BD. The two acidic regions involved in the binding to cytochrome P450 are also shown. TM: transmembrane region, BD: binding domain.
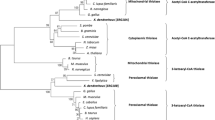
A phylogenetic comparison was performed using available fungal CPR protein sequences in the database by the Neighbor Joining method after 1000 replications of bootstrap analysis (Figure 4). The phylogenetic tree shows the evolutionary relationship of CrtR and allows the differentiation of three fungal clusters: Basidomycota, Ascomycota and Zygomycota. CrtR from X. dendrorhous is most closely related to the basidomycetes C. versicolor and to P. chrysosporium, with sequence identities of 60.2% and 58.3%, respectively.
Phylogenetic tree for cytochrome P450 reductases. Distance analysis and tree construction was done using the Neighbor Joining method. Numbers at nodes indicate the percent support for specific nodes after 1000 replications of bootstrap analysis. The scale bar indicates 10% change estimated sequence difference. The percentage of sequence identity with X. dendrorhous CrtR is in parenthesis. Accession numbers: Neurospora crassa: [GenBank:XP_964443]; Gibberella fujikuroi: [GenBank:AJ576025]; Cochliobolus lunatus: [GenBank:ABW86977]; Coccidioides immitis: [GenBank:XP_001246449]; Saccharomyces cerevisiae: [GenBank:D13788]; Candida tropicalis: [GenBank:M35199]; Rhodotorula minuta: [GenBank:AB055119]; Xanthophyllomyces dendrorhous: [GenBank:EU884134] and [GenBank:EU884133]; Coriolus versicolor: [GenBank:AB065368]; Phanerochaete chrysosporium: [GenBank:AF193060]; Cunninghamella elegans: [GenBank:AF195659]; Rhizopus stolonifer: [GenBank:AF290425]; Arabidopsis thaliana: [GenBank:A75959].
Expression of crtRgene
The relationship between the crtS and crtR gene expression along the growth curve at the mRNA level was studied in the wild type UCD 67–385 X. dendrorhous strain, grown in MMV supplemented with glucose as a fermentable carbon source. It was observed that crtS mRNA level reaches a maximum at 66 h of cultivation, equivalent to approximately half of the exponential growth phase. However, the levels of crtR mRNA remained constant along the growth curve (Figure 5). Despite the fact that both proteins are involved in the same stage of astaxanthin biosynthesis, there is no relationship, at the mRNA level, in their gene expression. It is possible to conclude that the two genes are not regulated in the same way at the transcriptional level.
According to this last result, in other work done in our laboratory, a dramatic decrease of transcripts levels from the crtYB, crtI and crtS genes (carotenogenic genes) was observed during the stationary phase [35]. This was not the case for the idi and crtE genes, where the level of the idi transcript showed a slight decline and the crtE transcript remained constant during the stationary phase. This suggests the presence of higher levels of the enzymes IPP isomerase and GGPP synthase at this stage. This situation would allow for the flow of the isoprenoid precursors towards carotenoid biosynthesis, as well as other metabolic pathways such as ergosterol biosynthesis [35]. Similarly, the absence of a relationship between the pattern of crtR gene expression and the rest of the carotenogenic genes suggests the existence of a different regulatory mechanism for crtR gene expression due to its involvement in other metabolic pathways of the yeast.
An example of cpr gene regulation has been described for Aspergillus niger. In this case, it was observed that the regulation of cpr gene expression was particularly complex in the P450-CPR system involved in benzoate hydroxylation [36]. In addition to benzoate induction at the transcriptional level, there were other regulatory mechanisms such as differential promoter use and post-translational regulatory mechanisms [36]. The decoupling of a P450-CPR system would imply the release of electrons to the cytoplasm that could generate active oxygen molecules, so a strict regulation of CPR is of vital importance for the cell. Similarly, it is not surprising that the crtS and crtR genes are regulated differently because, as mentioned earlier, several genes for different cytochrome P450 enzymes can exist in an organism, but generally there is a single gene for a unique cytochrome P450 reductase. Therefore, complex regulation for CrtR protein expression is necessary to adjust the levels of cytochrome P450 reductase activity in a cell with different levels of diverse cytochrome P450 proteins.
Functional study of the crtRgene
The participation of the crtR gene product in the formation of astaxanthin from β-carotene was studied in a bacterial heterologous system. For this, the DS1B E. coli strain (Table 1) was transformed with a pET101/D-TOPO expression vector carrying the cDNA of crtS and crtR genes from X. dendrorhous, and the carotenoid production was analyzed by RP-HPLC. This system was not successful, as it was not possible to produce any xanthophylls in the bacteria. A similar situation was observed in another study, where only canthaxanthin (β-carotene-4, 4'-dione, which is an intermediate in the conversion of β-carotene into astaxanthin) in very small quantities was produced when the S. cerevisiae cpr gene was used [12]. These authors concluded that the X. dendrorhous astaxanthin synthase has very low activity in E. coli [12]. It should be noted that the P450 systems are associated with membranes; therefore, E. coli may not provide a proper environment for its functionality. Recently, it was reported that several attempts to produce carotenoids in E. coli using the X. dendrorhous carotenogenic genes resulted in poor enzyme expression and carotenoid production [37]. Hence, a eukaryotic system would be more appropriate to attempt the expression of X. dendrorhous carotenogenic genes.
As the E. coli expression system was not successful, in order to demonstrate the importance of the crtR gene in the astaxanthin biosynthetic pathway, X. dendrorhous crtR deletion mutant strains were generated. The wild type strains UCD 67–385 and CBS-6938 were transformed with the linearized plasmid pBsiWIΔcrtR::hph and circular plasmid pNdeIΔcrtR::hph, respectively. Through homologous recombination events, the wild type crtR gene in the wild type strains was exchanged by the DNA fragment containing the deletion and resistance marker (Figure 6).
Graphical representation of transformation events. Wild type strains UCD 67–385 and CBS-6938 were transformed with the linearized plasmid pBsiWIΔcrtR::hph and circular plasmid pNdeIΔcrtR::hph, respectively. I, II, III: represent crtR exons 1, 2 and 3; P: EF-1α promoter, Term: GPDH transcription terminator; hph: hygromycin B resistance gene; pBS: pBluescript; BsiWI** and NdeI**: modified restriction sites after hygromycin B cassette insertion and arrows represent primer binding sites.
Through this experiment, a yellow (CBSTr, which derives from CBS-6938) and a pale red-orange (T13, which derives from UCD 67–385) hygromycin B resistant transformant strains were obtained. The color phenotype of these transformants is an indicator of an alteration in astaxanthin biosynthesis because astaxanthin has a strong red-orange color. The strain UCD 67–385 is diploid [30]; therefore, we suggest that T13 is heterozygote for the crtR gene, with a mutant and a wild type allele. Moreover, by a gene-dose effect, T13 produces less astaxanthin and accumulates more β-carotene than its parental wild type strain. On the other hand, the ploidy level of CBS-6938 is unknown. However, based on random mutagenesis results with physical and chemical mutagens performed in our laboratory (data not shown) and transformation with carotenogenic genes [38], it was concluded that this strain could be haploid. The mutagenesis results suggest that, in the case of CBSTr, the only crtR gene copy is mutated and, therefore, it is not capable of producing astaxanthin and accumulates β-carotene.
In order to confirm the T13 (crtRΔBsiWI::hph/crtR+) and CBSTr (crtRΔNdeI::hph) transformant genotype modifications, PCR reactions were performed using specific primers for the crtR and hph genes (Table 2), and genomic DNA samples from wild type and transformant strains were used as templates. The amplicon sequences validated the representation shown in Figure 6. The results indicated that the CBS-6938 strain has one crtR gene copy and its deletion is not lethal. In the case of T13, it was shown that it is heterozygote for the crtR gene, having a wild type and a mutant allele, which supports the gene-dose effect hypothesis in the production of astaxanthin. These results were also corroborated by Southern blot hybridization (data not shown).
Several attempts were performed to obtain a β-carotene accumulating transformant derived from UCD 67–385. T13 was retransformed with a DNA fragment harboring a crtR gene deletion and a G418 resistance cassette marker. No G418 resistant transformants were obtained. In order to produce a homozygous crtR mutation, T13 was subjected to the Double Recombinant Method [6], but it was not possible to obtain a double mutant strain for the crtR gene.
The cpr gene deletion in S. cerevisiae was not lethal, suggesting the existence of an alternative electron donor such as a cytochrome b5 (cytb5) [39]. When a cytb5 deletion in the wild type was done, no new phenotype was generated. However, it was lethal when cytb5 and cpr were simultaneously disrupted. This result demonstrated that, in mutants with single disruptions of cpr or cytb5, both enzymes can complement each other [39]. Likewise, the disruption of the cpr gene from the fungus Gibberella fujikuroi was not lethal, proving the existence of another electron donor [16]. In light of these results and evidence for the existence of a high level of polymorphisms in different X. dendrorhous strains [21, 30, 31], we suggest that both wild type strains may have a different genetic background. Moreover, the results obtained in this work suggest that, in the CBS-6938 wild type strain, an alternative electron donor such as a cytochrome b5 may exist because the crtR gene disruption was not lethal. On the other hand, this is not the case for the UCD 67–385 strain due to the impossibility to obtain a homozygous mutant for the crtR gene. However, the possible alternative electron donor in the CBS-6938 strain cannot support astaxanthin synthesis, as the crtR gene mutant (CBSTr) accumulates β-carotene.
To check the carotenoid composition, total pigments were extracted from both wild type and transformant strains, and the composition was analyzed by RP-HPLC (Figure 7 and Table 4). The RP-HPLC analysis showed that the carotenoid composition is completely different between transformants and their parental strain. CBSTr accumulates β-carotene and is unable to synthesize astaxanthin. T13 has a reduced astaxanthin production at the expense of increased β-carotene production, which is the astaxanthin precursor.
RP-HPLC analysis of carotenoids from wild type (UCD 67–385, CBS-6983) and transformant (T13 ( crtRΔBsiWI::hph/ crtR+), CBSTr ( crtRΔNdeI::hph)) strains. Numbers indicate different carotenoids as follows: 1: astaxanthin; 2: phoenicoxanthin; 3: canthaxanthin; 4: hydroxy-keto-γ-carotene; 5: keto-γ-carotene; 6: hydroxy-echinenone; 7: echinenone; 8: lycopene; 9: neurosporene; 10: γ-carotene and 11: β-carotene. In the upper right of the chromatogram, colony phenotype is shown.
Conclusion
A new crtR gene has been isolated and characterized from X. dendrorhous, which plays an auxiliary role to the crtS gene and is also essential for astaxanthin biosynthesis. This gene consists of three exons, and is localized in at least two chromosomes of the UCD 67–385 X. dendrorhous wild type strain. crtR encodes a 746 amino acid protein with characteristic domains of cytochrome P450 reductases (FMN, FAD and NAD(P)H binding domains) and a small amino terminal transmembrane region. Although both crtS and crtR proteins participate in the same stage of yeast carotenogenesis, the mRNA level pattern from these genes along the growth curve is different. Thus, both genes are regulated in a different manner at the transcriptional level.
References
Takaichi S, Sandmann G, Schnurr G, Satomi Y, Suzuki A, Misawa N: The carotenoid 7,8-dihydro-ψ end group can be cyclized by the licopene cyclases from the bacterium Erwinia uredovora and the higher plant Capsicum annuum. Eur J Biochem. 1996, 241: 291-296. 10.1111/j.1432-1033.1996.0291t.x.
Johnson E, Conklin D, Lewis M: The yeast Phaffia rhodozyma as a dietary pigment source for salmonids and crustaceans. J Fish Res Board Can. 1997, 34: 2417-2421.
Andrewes AG, Phaff HJ, Starr MP: Carotenoids of Phaffia rhodozyma. A red pigmented fermenting yeast. Phytochem. 1976, 15: 1003-1007. 10.1016/S0031-9422(00)84390-3.
Fraser P, Miura Y, Misawa N: In vitro characterization of astaxanthin biosynthetic enzymes. J Biol Chem. 1997, 272: 6128-6135. 10.1074/jbc.272.10.6128.
Kajiwara S, Fraser PD, Kondo K, Misawa N: Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem J. 1997, 324: 421-426.
Niklitschek M, Alcaíno J, Barahona S, Sepúlveda D, Carmona M, Wozniak A, Marcoleta A, Lodato P, Baeza M, Cifuentes V: Genomic organization of the structural gene controlling the astaxanthin biosynthesis pathway of Xanthophyllomyces dendrorhous. Biol Res. 2008, 41: 93-108.
Verdoes JC, Krubasik P, Sandmann G, van Ooyen AJJ: Isolation and funtional characterisation of a novel type of carotenoid biosynthetic gene from Xanthophyllomyces dendrorhous. Mol Gen Genet. 1999, 262: 453-461. 10.1007/s004380051105.
Verdoes JC, Misawa N, van Ooyen AJJ: Cloning and characterization of the astaxanthin biosynthetic gene encoding phytoene desaturase of Xanthophyllomyces dendrorhous. Biotechnol Bioeng. 1999, 63: 750-755. 10.1002/(SICI)1097-0290(19990620)63:6<750::AID-BIT13>3.0.CO;2-7.
Cunningham FX, Gantt E: Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol. 1998, 49: 557-583. 10.1146/annurev.arplant.49.1.557.
Linden H: Carotenoid hydroxylase from Haematococcus pluvialis: cDNA sequence, regulation and functional complementation. Biochim Biophys Acta. 1999, 1446: 203-212.
Hoshino T, Ojima K, Setoguchi Y: Astaxanthin synthase. Hoffman-LaRoche, European patent no.1035206-A3. 2000
Ojima K, Breitenbach J, Visser H, Setoguchi Y, Tabata K, Hoshino T, Berg van den J, Sandmann G: Cloning of the astaxanthin synthase gene from Xanthophyllomyces dendrorhous (Phaffia rhodozyma) and its assignment as a beta-carotene 3-hydroxylase/4-ketolase. Mol Genet Genomics. 2006, 275: 148-158. 10.1007/s00438-005-0072-x.
Alvarez V, Rodriguez-Saiz M, de la Fuente JL, Gudina EJ, Godio RP, Martín JF, Barredo JL: The crtS gene of Xanthophyllomyces dendrorhous encodes a novel cytochrome-P450 hydroxylase involved in the conversion of beta-carotene into astaxanthin and other xanthophylls. Fungal Genet Biol. 2006, 43: 261-272. 10.1016/j.fgb.2005.12.004.
Martín JF, Gudiña E, Barredo JL: Conversion of beta-carotene into astaxanthin: Two separate enzymes or a bifunctional hydroxylase-ketolase protein?. Microb Cell Fact. 2008, 20: 7-13.
van den Brink HM, van Gorcom RF, van den Hondel CA, Punt PJ: Cytochrome P450 enzyme systems in fungi. Fungal Genet Biol. 1998, 23: 1-17. 10.1006/fgbi.1997.1021.
Malonek S, Rojas MC, Hedden P, Gaskin P, Hopkins P, Tudzynski B: The NADPH-cytochrome P450 reductase gene from Gibberella fujikuroi is essential for gibberellin biosynthesis. J Biol Chem. 2004, 279: 25075-25084. 10.1074/jbc.M308517200.
Retamales P, Hermosilla G, León R, Martínez C, Jiménez A, Cifuentes V: Development of the sexual reproductive cycle of Xanthophyllomyces dendrorhous. J Microbiol Methods. 2002, 48: 87-93. 10.1016/S0167-7012(01)00349-9.
Sambroock J, Russell DW: Molecular cloning. A laboratory manual. 2001, Cold Spring Harbor NY: Cold Spring Harbor Laboratory Press, 3
Guengerich FP, Martin MV, Guo Z, Chun YJ: Purification of functional recombinant P450s from bacteria. Methods Enzymol. 1996, 272: 35-44. 10.1016/S0076-6879(96)72006-2.
Boyle JS, Lew AM: An inexpensive alternative to glassmilk for DNA purification. TIG. 1995, 11: 8-
Cifuentes V, Hermosilla G, Martínez C, León R, Pincheira G, Jiménez A: Genetics and electrophoretic karyotyping of wild type and astaxantin mutants strains from Phaffia rhodozyma. Antonie Van Leeuwenhoek. 1997, 72: 111-117. 10.1023/A:1000200119447.
Chomczynski P, Sacchi YN: Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987, 162: 156-159. 10.1016/0003-2697(87)90021-2.
Lodato P, Alcaíno J, Barahona S, Retamales P, Jiménez A, Cifuentes V: Study of the expresion of carotenoid biosintesis gene in wild type and deregulated strains of Xanthophyllomyces dendrorhous (Ex Phaffia rhodozyma). Biol Res. 2004, 37: 83-93.
Wery J, Dalderup MJ, Ter Linde J, Boekhout T, van Ooyen AJJ: Structural and phylogenetic analysis of the actin gene from the yeast Phaffia rhodozyma. Yeast. 1996, 12: 641-651. 10.1002/(SICI)1097-0061(19960615)12:7<641::AID-YEA952>3.0.CO;2-2.
Ngiam C, Jeenes DJ, Punt PJ, Hondel van den CA, Archer DB: Characterization of a foldase, protein disulfide isomerase A, in the protein secretory pathway of Aspergillus niger. Appl Environ Microbiol. 2000, 66: 775-782. 10.1128/AEM.66.2.775-782.2000.
Adrio J, Veiga M: Transformation of the astaxanthin-producing yeast Phaffia rhodozyma. Biotech Techniques. 1995, 9: 509-512. 10.1007/BF00159567.
Kim IG, Nam SK, Sohn JH, Rhee SK, An GH, Lee SH, Choi ES: Cloning of the ribosomal protein L41 gene of Phaffia rhodozyma and its use as a drug resistance marker for transformation. Appl Environ Microbiol. 1998, 64: 1947-1949.
Reyes E, Barahona S, Fischman O, Niklitschek M, Baeza M, Cifuentes V: Genetic polymorphism of clinical and environmental strains of Pichia anomala. Biol Res. 2004, 37: 747-57.
An GH, Schuman DB, Johnson EA: Isolation of Phaffia rhodozyma mutants with increased astaxanthin content. Appl Environ Microbiol. 1989, 55: 116-124.
Hermosilla G, Martínez C, Retamales P, León R, Cifuentes V: Genetic determination of ploidy level in Xanthophyllomyces dendrorhous. Antonie van Leeuwenhoek. 2003, 84: 279-287. 10.1023/A:1026090008405.
Nagy A, Garamszegib N, Vágvölgyib C, Ferenczyb L: Electrophoretic karyotypes of Phaffia rhodozyma strains. FEMS Microbiol Lett. 1994, 123: 315-318. 10.1111/j.1574-6968.1994.tb07241.x.
Yadav JS, Loper JC: Cytochrome P450 oxidoreductase gene and its differentially terminated cDNAs from the white rot fungus Phanerochaete chrysosporium. Curr Genet. 2000, 37: 65-73. 10.1007/s002940050010.
Porter TD, Kasper CB: NADPH-cytochrome P-450 oxidoreductase: flavin mononucleotide and flavin adenine dinucleotide domains evolved from different flavoproteins. Biochem. 1986, 25: 1682-1687. 10.1021/bi00355a036.
Shen AL, Kasper CB: Role of acidic residues in the interaction of NADPH-cytochrome P450 oxidoreductase with cytochrome P450 and cytochrome c. J Biol Chem. 1995, 270: 27475-27480. 10.1074/jbc.270.46.27475.
Lodato P, Alcaíno J, Barahona S, Niklitschek M, Carmona M, Wozniak A, Baeza M, Jiménez A, Cifuentes V: Expression of the carotenoid biosynthesis gene in Xanthophyllomyces dendrorhous. Biol Res. 2007, 40: 73-84.
van den Brink JM, Punt PJ, van Gorcom RF, van den Hondel CA: Regulation of expression of the Aspergillus niger benzoate para-hydroxylase cytochrome P450 system. Mol Gen Genet. 2000, 263: 601-609. 10.1007/s004380051207.
Verwaal R, Wang J, Meijnen JP, Visser H, Sandmann G, Berg van den JA, van Ooyen AJJ: High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous. Appl Environ Microbiol. 2007, 73: 4342-4350. 10.1128/AEM.02759-06.
Verdoes JC, Sandmann G, Visser H, Diaz M, van Mossel M, van Ooyen AJJ: Metabolic Engineering of the Carotenoid Biosynthetic Pathway in the Yeast Xanthophyllomyces dendrorhous (Phaffia rhodozyma). Appl Environ Microbiol. 2003, 69 (7): 3728-38. 10.1128/AEM.69.7.3728-3738.2003.
Truan G, Epinat JC, Rougeulle C, Cullin C, Pompon D: Cloning and characterization of a yeast cytochrome b5-encoding gene which suppresses ketoconazole hypersensitivity in a NADPH-P-450 reductase-deficient strain. Gene. 1994, 142: 123-127. 10.1016/0378-1119(94)90366-2.
Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K: Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol. 1990, 172: 6704-6712.
Acknowledgements
This work was supported by U. de Chile ENL 08/06 and: MECESUP UCH 0106 by graduate scholarships to JA and MN, Fundación María Ghilardi by a graduate scholarship to CL, CONICYT by a graduate scholarship to AM.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
JA participated in the design of this study and carried out the crtR gene isolation, sequence analysis, crtR gene expression study, X. dendrorhous transformation, pigment extraction, RP-HPLC and drafted the manuscript. SB constructed the X. dendrorhous YIp5 genomic library. MC participated in the isolation of the crtS gene genomic and cDNA versions. CL participated in the UCD 67–385 strain fermentation and RT-PCR analysis. AM constructed the X. dendrorhous cDNA library. MN constructed the hygromycin B resistance cassette. DS participated in DNA sequencing. MB participated in the study design. VC conceived the study and participated in its design and coordination. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Alcaíno, J., Barahona, S., Carmona, M. et al. Cloning of the cytochrome p450 reductase (crtR) gene and its involvement in the astaxanthin biosynthesis of Xanthophyllomyces dendrorhous. BMC Microbiol 8, 169 (2008). https://doi.org/10.1186/1471-2180-8-169
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-8-169