Abstract
Background
Increasingly, metabolite and reaction information is organized in the form of genome-scale metabolic reconstructions that describe the reaction stoichiometry, directionality, and gene to protein to reaction associations. A key bottleneck in the pace of reconstruction of new, high-quality metabolic models is the inability to directly make use of metabolite/reaction information from biological databases or other models due to incompatibilities in content representation (i.e., metabolites with multiple names across databases and models), stoichiometric errors such as elemental or charge imbalances, and incomplete atomistic detail (e.g., use of generic R-group or non-explicit specification of stereo-specificity).
Description
MetRxn is a knowledgebase that includes standardized metabolite and reaction descriptions by integrating information from BRENDA, KEGG, MetaCyc, Reactome.org and 44 metabolic models into a single unified data set. All metabolite entries have matched synonyms, resolved protonation states, and are linked to unique structures. All reaction entries are elementally and charge balanced. This is accomplished through the use of a workflow of lexicographic, phonetic, and structural comparison algorithms. MetRxn allows for the download of standardized versions of existing genome-scale metabolic models and the use of metabolic information for the rapid reconstruction of new ones.
Conclusions
The standardization in description allows for the direct comparison of the metabolite and reaction content between metabolic models and databases and the exhaustive prospecting of pathways for biotechnological production. This ever-growing dataset currently consists of over 76,000 metabolites participating in more than 72,000 reactions (including unresolved entries). MetRxn is hosted on a web-based platform that uses relational database models (MySQL).
Similar content being viewed by others
Background
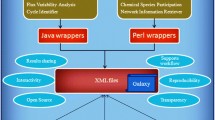
The ever accelerating pace of DNA sequencing and annotation information generation [1] is spearheading the global inventorying of metabolic functions across all kingdoms of life. Increasingly, metabolite and reaction information is organized in the form of community [2], organism, or even tissue-specific genome-scale metabolic reconstructions. These reconstructions account for reaction stoichiometry and directionality, gene to protein to reaction associations, organelle reaction localization, transporter information, transcriptional regulation and biomass composition. Already over 75 genome-scale models are in place for eukaryotic, prokaryotic and archaeal species [3] and are becoming indispensable for computationally driving engineering interventions in microbial strains for targeted overproductions [4–7], elucidating the organizing principles of metabolism [8–11] and even pinpointing drug targets [12, 13]. A key bottleneck in the pace of reconstruction of new high quality metabolic models is our inability to directly make use of metabolite/reaction information from biological databases [14] (e.g., BRENDA [15], KEGG [16], MetaCyc, EcoCyc, BioCyc [17], BKM-react [18], UM-BBD [19], Reactome.org, Rhea, PubChem, ChEBI etc.) or other models [20] due to incompatibilities of representation, duplications and errors, as illustrated in Figure 1.
A major impediment is the presence of metabolites with multiple names across databases and models, and in some cases within the same resource, which significantly slows down the pooling of information from multiple sources. Therefore, the almost unavoidable inclusion of multiple replicates of the same metabolite can lead to missed opportunities to reveal (synthetic) lethal gene deletions, repair network gaps and quantify metabolic flows. Moreover, most data sources inadvertently include some reactions that may be stoichiometrically inconsistent [21] and/or elementally/charge unbalanced [22, 23], which can adversely affect the prediction quality of the resulting models if used directly. Finally, a large number of metabolites in reactions are partly specified with respect to structural information and may contain generic side groups (e.g., alkyl groups -R), varying degree of a repeat unit participation in oligomers, or even just compound class identification such as "an amino acid" or "electron acceptor". Over 3% of all metabolites and 8% of all reactions in the aforementioned databases and models exhibit one or more of these problems.
There have already been a number of efforts aimed at addressing some of these limitations. The Rhea database, hosted by the European Bioinformatics Institute, aggregates reaction data primarily from IntEnz [24] and ENZYME [25], whereas Reactome.org is a collection of reactions primarily focused on human metabolism [26, 27]. Even though they crosslink their data to one or more popular databases such as KEGG, ChEBI, NCBI, Ensembl, Uniprot, etc., both retain their own representation formats. More recently, the BKM-react database is a non-redundant biochemical reaction database containing known enzyme-catalyzed reactions compiled from BRENDA, KEGG, and MetaCyc [18]. The BKM-react database currently contains 20,358 reactions. Additionally, the contents of five frequently used human metabolic pathway databases have been compared [28]. An important step forward for models was the BiGG database, which includes seven genome-scale models from the Palsson group in a consistent nomenclature and exportable in SBML format [29–31]. Research towards integrating genome-scale metabolic models with large databases has so far been even more limited. Notable exceptions include the partial reconciliation of the latest E. coli genome scale model i AF1260 with EcoCyc [32] and the aggregation of data from the Arabidopsis thaliana database and KEGG for generating genome-scale models [33] in a semi-automated fashion. Additionally, ReMatch integrates some metabolic models, although its primary focus is on carbon mappings for metabolic flux analysis [34]. Also, many metabolic models retain the KEGG identifiers of metabolites and reactions extracted during their construction [35, 36]. An important recent development is the web resource Model SEED that can generate draft genome-scale metabolic models drawing from an internal database that integrates KEGG with 13 genome scale models (including six of the models in the BiGG database) [37]. All of the reactions in Model SEED and BiGG are charge and elementally balanced.
In this paper, we describe the development and highlight applications of the web-based resource MetRxn that integrates, using internally consistent descriptions, metabolite and reaction information from 8 databases and 44 metabolic models. The MetRxn knowledgebase (as of October 2011) contains over 76,000 metabolites and 72,000 reactions (including unresolved entries) that are charge and elementally balanced. By conforming to standardized metabolite and reaction descriptions, MetRxn enables users to efficiently perform queries and comparisons across models and/or databases. For example, common metabolites and/or reactions between models and databases can rapidly be generated along with connected paths that link source to target metabolites. MetRxn supports export of models in SBML format. New models are being added as they are published or made available to us. It is available as a web-based resource at http://metrxn.che.psu.edu.
Construction and Content
MetRxn construction
The construction of MetRxn largely followed the following steps, as illustrated in Figure 2: 1) download of primary sources of data from databases and models, 2) integration of metabolite and reaction data, 3) calculation and reconciliation of structural information, 4) identification of overlaps between metabolite and reaction information, 5) elemental and charge balancing of reactions, 6) successive resolution of remaining ambiguities in description.
Flowchart outlining the construction of MetRxn. After download of primary sources of data from databases and models, we integrated metabolite and reaction data, followed by calculation and reconciliation of structural information. By identifying overlaps between metabolite and reaction information, we generated elemental and charge balancing of reactions. The procedure for developing MetRxn was iterative with subsequent passes making use of previous associations to resolve remaining ambiguities.
Step 1: Source data acquisition
Metabolite and reaction data was downloaded from BRENDA, KEGG, BioCyc, BKM-react and other databases using a variety of methods based on protocols such as SOAP, FTP and HTTP. We preprocessed the data into flat files that were subsequently imported into the knowledgebase. All original information pertaining to metabolite name, abbreviations, metabolite geometry, related reactions, catalyzing enzyme and organism name, gene-protein-reaction associations, and compartmentalization was retained. For all 44 initial genome-scale models listed, the online information from the corresponding publications was also imported. The source codes for all parsers used in Step 1 are available on the MetRxn website.
Step 2: Source data parsing
The "raw data" from both databases and models was unified using standard SQL scripts on a MySQL server. The description schema for metabolites includes source, name, abbreviations used in the source, chemical formula, and geometry. The schema for reactions accounts for source, name, reaction string (reactants and products), organism designation, associated enzymes and genes, EC number, compartment, reversibility/direction, and pathway information. Once a source has been imported into the MySQL server, a data source-specific dictionary is created to map metabolite abbreviations onto names/synonyms and structures and metabolites to reactions.
Step 3: Metabolite charge and structural analysis
We used Marvin (Chemaxon) to analyze all 218,122 raw metabolite entries containing structural information (out of a total of 322,936, including BRENDA entries). Inconsistencies were found in 12,965 entries typically due to wrong atom connectivity, valence, bond length or stereo chemical information, which were corrected using APIs available in Marvin. A final corrected version of the metabolite geometries was calculated at a fixed pH of 7.2 and converted into standard Isomeric SMILES format. The structure/formula used corresponded to the major microspecies found during the charge calculation, which effectively rounds the charge to an integer value in accordance with previous model construction conventions. This format includes both chiral and stereo information, as it allows specification of molecular configuration [38–40]. Metabolites were also annotated with Canonical SMILES using the OpenBabel Interface from Chemspider. The canonical representation encodes only atom-atom connectivity while ignoring all conformers for a metabolite. Using bond connectivity information from the primary sources and resources such as PubChem and ChemSpider we used Canonical SMILES [41, 42] to resolve the identity of 34,984 metabolites and 32,311 reactions. Another 6,100 metabolites and 11,401 reactions involved, in various degrees, lack of full atomistic detail in their description (e.g., use an R or × as side-chains, are generic compounds like "amino acid" or "electron acceptor"). Over 25,000 duplicate metabolites and 27,000 reaction entries were identified and consolidated within the database. The metabolites and reactions present in the resolved repository were further classified with respect to the completeness of atomistic detail in their description.
Step 4: Metabolite synonyms and initial reaction reconciliation
Raw metabolite entries were assigned to Isomeric SMILES representations whenever possible. If insufficient structural information was available for a downloaded raw metabolite then it was assigned temporarily with the Canonical SMILES and revisited during the reaction reconciliation. Canonical SMILES retain atom connectivity but not stereo-specificity and are used as the basic metabolite topology descriptors as many metabolic models lack stereo-specificity information. After generating the initial metabolite associations, we identified reaction overlaps using the reaction synonyms and reaction strings along with the metabolite SMILES representations. Directionality and cofactor usage were temporarily ignored. During this step, reactions were flagged as single-compartment or two-compartment (i.e., transport reactions). MetRxn internally retains the original compartment designations, but currently only displays these simplified compartment designations. In analogy to metabolites, reactions were grouped into families that shared participants but in the source data sets occurred in different compartments or differed only in protonation.
Step 5: Reaction charge and elemental balancing
Once metabolites were assigned correct elemental composition and protonation states, reactions were charge and elementally balanced. To this end, for charge balancing we relied on a linear programming representation that minimizes the difference in the sum of the charge of the reactants and the sum of the charge on the products. The complete formulation is provided in the documentation at MetRxn.
Step 6: Iterative reaction reconciliation
Reactions with one (or more) unresolved reactants and/or products were string compared against the entire resolved collection of reactions. This step was successively executed as newly resolved metabolites and reactions could enable the resolution of previously unresolved ones. After the first pass 164 metabolites were resolved, while subsequent passes (up to 18 for some models) helped resolved a total of 8,720 entries. Reactions with significant (but not complete) overlapping sets of reactants/products are additionally sent to the curator GUI including phonetic information. Briefly, the phonetic tokens of synonyms with known structures were compared against the ones without any associated structure. The algorithm suppresses keywords/tokens depicting stereo information such as cis, trans, L-, D-, alpha, beta, gamma, and numerical entries because they change the phonetic signature of the synonym under investigation. In addition, the algorithm ignores non-chemistry related words (e.g., use, for, experiment) that are found in some metabolite names. Certain tokens such as "-ic acid" and "-ate" are treated as equivalent. PubChem and Chemspider sources were accessed through the GUI so that the curator gets as much information as possible to identify the data correctly. Phonetic matches provided clues for resolving over 159 metabolites. The iterative application of string and phonetic comparison algorithms resolved as many as 8,879 metabolites after 18 rounds of reconciliation.
Upon completion of this workflow, all genome-scale models are reformatted into a computations-ready form and Flux Balance Analysis [43] is performed on both the source model and the standardized model in MetRxn to ascertain the ability of the model to produce biomass before and after standardization. We performed the calculations using GAMS version 12.6. MetRxn is accessible through a web interface that indirectly generates MySQL queries. In order to facilitate analysis and use of the data, a number of tools are provided as part of MetRxn.
Data export and display
MetRxn supports a number of export capabilities. In general, any list that is displayed contains live links to the metabolite or reaction entities. These lists can consist of an entire model, data from a comparison, or query results. All items can be exported to SMBL format. In addition, the public MySQL database will be made available upon request. Because of licensing limitations, the BRENDA database cannot be exported and is not part of the public MySQL database. However, we plan to provide Java source code that allows for the integration of a local copy of the public MySQL database with the BRENDA database (provided upon request).
Source comparisons and visualization
In addition to listing the content (number of metabolites, reactions, etc.) of the selected data source(s), MetRxn contains tools for comparing two or more models and visualizing the results. These associations can be for metabolites or reactions. During these comparisons compartment information and reversibility are suppressed. Comparison tables are generated by comparing the associations between the selected data source(s) using the canonical structures.
MetRxn Scope
An initial repository of reaction (i.e., 154,399) and metabolite (i.e., 322,936) entries were downloaded from 8 databases and 44 genome-scale metabolic models. We compiled a non-redundant list of 42,540 metabolites and 35,474 reactions (after consolidating duplicate entries) containing full atomistic and bond connectivity detail. Another 6,100 metabolites and 11,401 reactions have partial atomistic detail typically containing generic side-chains (R) and/or an unspecified number of polymer repeat units. Finally, 5,436 metabolites in metabolic models and 8,000 metabolites in databases are retained with no atomistic detail. In some cases lack of atomistic detail reflects complete lack of identity specificity (e.g., electron donor) whereas in other cases even though the chemical species is fully defined, atomistic level description is not warranted (e.g., gene product of dsbC protein disulfide isomerase II (reduced)). Figure 3 shows the distribution of metabolite resolution across models and databases in MetRxn. In general, metabolites without fully-specified structures tend to participate in a relatively small number of reactions.
Various levels of structural information was available for models (main) and databases (inset). For every model, the majority of metabolites had full atomistic detail (blue). The smaller number of metabolites with partial atomistic detail (orange) such as genetic side chains, or with no atomistic detail (green) such as gene products, participated in few reactions.
The workflow followed in the creation of the MetRxn knowledgebase identified a number of inconsistencies. For instance, the same metabolite name may map to molecules with different numbers of repeat units (e.g., lecithin) or completely different structures (e.g., AMP could refer to either adenosine monosphate or ampicillin). Notably, even for the most well-curated metabolic model, E. coli i AF1260 [32], we found minor errors or omissions (a total of 17) arising from inconsistencies or incompleteness of representation in the data culled from other sources. For example, the metabolite abbreviation arbtn-fe3 was mistakenly associated with the KEGG ID and structure of aerobactin instead of ferric-aerobactin. The number of inconsistencies is dramatically increased for less-curated metabolic models. We used a variety of procedures to disambiguate the identity of metabolites lacking structural information ranging from reaction matching to phonetic searches. For example, in the Corynebacterium glutamicum model [44], 7,8-aminopelargonic acid (DAPA) has no associated structural information. Reaction matching found the same reaction in the E. coli i AF1260 model.
which implies that 7,8-aminopelargonic acid (DAPA) is identical to 7,8-Diaminononanoate (dann). Examination of pelargonic acid and nonanoate reveals that they were indeed known synonyms. In many cases, we were also able to assign stereo-specific information to metabolite entries in models (e.g., stipulate the L-lysine isomer for lysine). We made use of an iterative approach that allowed us to map structures from models with explicit links to structures (e.g. to KEGG or CAS numbers) to models that only provided metabolite names. Furthermore, by using a phonetic algorithm that uses tokens for equivalent strings in metabolite names (e.g., '-ic acid' and '-ate' are equivalent) we were able to resolve more than an additional 159 metabolites. For example, phonetic searches flagged cis-4-coumarate and COUMARATE in the Acinetobacter baylyi model [45] as potentially identical compounds. Additional checks revealed that indeed both metabolites should map to the same structure. A more complex matching example involved 1-(5'-Phosphoribosyl)-4-(N-succinocarboxamide)-5-aminoimidazole from the Bacillus subtilis model [46] and 1-(5'-Phosphoribosyl)-5-amino-4-(N-succinocarboxamide)-imidazole from the Aspergillus nidulans model [47]. We note that the phonetic algorithm only makes suggestions and orders the possible matches for the curator. Next, we detail three examples that provide an insight into the type of tasks that MetRxn can facilitate.
Utility and Discussion
1. Charge and elementally balanced metabolic models
The standardized description of metabolites and balanced reactions afforded by MetRxn enables the expedient repair of existing models for metabolite naming inconsistencies and reaction balancing errors. Here we highlight one such metabolic model repair for Acinetobacter baylyi i Abaylyiv4 [45]. We identified that 189 out of 880 reactions are not elementally or charge balanced. Most of the reactions with charge balance errors involved a missed proton in reactions involving cofactor pairs such as NAD/NADH. For example, a proton had to be added to the reactants side in the reaction (R, R)-Butanediol-dehydrogenase in which butanediol reacts with NAD to form acetoin. In addition, the stoichiometric coefficient of water in GTP cyclohydrolase I was erroneously set at -2 which resulted in an imbalance in oxygen atoms. The re-balancing analysis changed the coefficient to -1 (as listed in BRENDA) and added a proton to the list of reactants (absent from BRENDA) in order to also balance charges.
We performed flux balance analysis (FBA) on both the published and MetRxn-based rebalanced version of the Acinetobacter baylyi model using the uptake constraints listed in [45] to assess the effect of re-balancing reaction entries on FBA results. We found that the maximum biomass using the glucose/ammonia uptake environment decreased by 9% primarily due to the increased energetic costs associated with maintaining the proton gradient. This result demonstrates the significant effect that lack of reaction balancing may cause in FBA calculations. Overall, we found that nearly two-thirds of the models had at least one unbalanced reaction, with over 2,400 entities across all models that were either charge or elementally imbalanced. Frequently, the same reaction was imbalanced in multiple models (each occurrence was counted separately).
2. Contrasting existing metabolic models
At the onset of creating MetRxn, we conducted a brief preliminary study to quantify the extent/severity of naming inconsistencies by contrasting the reaction information contained in an initial collection of 34 of the most popular genome-scale models spanning 21 bacterial, 10 eukaryotic and three archaeal organisms. Across all branches of life, most metabolic processes are largely conserved (e.g., glycolysis, pentose phosphate pathway, amino acid biosynthesis, etc.) therefore we expected to uncover a large core of common reactions shared by all models. Surprisingly, we found that only three reactions (i.e., phosphoglycerate mutase, phosphoglycerate kinase, and CO2 transport) were directly recognized as common across those 34 models using a simple string match comparison. Even when examining models for only a few bacterial organisms (Bacillus subtilis, Escherichia coli, Mycobacterium tuberculosis, Mycoplasma genitalium, and Salmonella Typhimurium) simple text searches recognized only 40 common reactions (out of a possible 262, which is the size of the M. genitalium model). The reason for this glaring inconsistency is that differing metabolite naming conventions, compartment designations, stoichiometric ratios, reversibility, and water/proton balancing issues prevents the automated recognition of genuinely shared reactions across models. Using the glucose-6-phosphate dehydrogenase reaction as a representative example, Table 1 reveals some of the reasons for failing to automatically recognize common reactions across selected models [13, 32, 35, 36, 47–64]. As many as nine different representations of the same reaction exist due to incomplete elemental and charge balancing, alternate cofactor usage among different organisms, and lack of universal metabolite naming conventions. We have found that this level of discord between models is representative for most metabolic reactions. This lack of consistency renders direct pathway comparisons across models meaningless and the aggregation of reaction information from multiple models precarious. This deficiency motivated the development of MetRxn. Given standardization in metabolite naming and elementally/charge balanced reaction entries MetRxn allows for the identification of shared reactions as well as differences between any two metabolic models (assuming that all the metabolites in the compared reaction entries have full atomistic information). When making the comparison of those same metabolic models, MetRxn found an additional 15 reactions in common (for a total of 55 -- a 38% increase) and that 142 reactions are shared by B. subtilis, E. coli and Salmonella Typhimurium.
The Web interface of MetRxn allows for any number of models to be simultaneously compared. As a demonstration of this capability we selected to contrast the metabolic content of two clostridia models: Clostridium acetobutylicum [65] and Clostridium thermocellum [66]. Figure 4 shows the results in the form of a Venn diagram. Some of the differences between the clostridia species are not surprising arising due to their differing lifestyles (C. acetobutylicum contains solventogenesis pathways and a CoB12 pathway, whereas C. thermocellum contains cellulosome reactions). However, we found many differences that appear to reflect different conventions adopted when the two models were generated rather than genuine differences in metabolism. In particular, in the C. thermocellum model [66] charged/uncharged tRNA metabolites are explicitly tracked whereas they are not included in the C. acetobutylicum model [65]. Surprisingly, both clostridia models are more similar, at the metabolite level, to the Bacillus subtilis iBsu1103 model [46] rather than to each other (see Figure 4). Charged/uncharged tRNA metabolites account for most of the increased overlap between C. thermocellum and B. subtilis. Most of the reaction overlaps are in the amino acids biosynthesis pathways, carbohydrate metabolism, and nucleoside metabolism. It is important to note that 48 reactions in C. acetobutylicum, 67 reactions in C. thermocellum, and 120 reactions in B. subtilis lack full atomistic information (see Figure 3) and thus were excluded from any comparisons. It is possible that additional shared reactions between the two models can be deduced by further examining comparisons between not fully structurally specified metabolite entries. The string/phonetic comparison algorithms described under Step 6 along with assisted curation could be adapted for this task.
Comparison of metabolite and reaction overlaps for C. acetobutylicum and C. thermocellum , and B. subtilis. Although the two Clostridium organisms are same genus, the models of these two species had significant numbers of unique metabolites (left) and reactions (right), and comparisons revealed that there was more similarity in metabolite usage with a model of B. subtilis than with each other. In part, these overlaps were driven by the explicit accounting for charged tRNA species in C. thermocellum and B. subtilis models, which was also reflected in the reaction overlaps through reactions involving these metabolites.
3. Using MetRxn to Bio-Prospect for Novel Production Routes
A "Grand Challenge" in biotechnological production is the identification of novel production routes that allow for the conversion of inexpensive resources (e.g., various sugars) into useful products (e.g., succinate, artemisinin) and bio-fuels (e.g., ethanol, butanol, biodiesel etc.). Selected production routes must exhibit high yields, avoid thermodynamic barriers, bypass toxic intermediates and circumvent existing intellectual property restrictions. Historically, the incorporation of heterologous pathways relied largely on human intuition and literature review followed by experimentation [67, 68]. Currently, rapidly expanding compilations of biotransformations such as KEGG [69] and BRENDA [70] are increasingly being prospected using search algorithms to identify biosynthetic routes to important product molecules. Several optimization and graph-based methods have been employed to computationally assemble novel biochemical routes from these sources. OptStrain [71] used a mixed-integer linear optimization representation to identify the minimal number of reactions to be added (i.e. knock-ins) into a genome-scale metabolic model to enable the production of the new molecule. However the combinatorial nature of the problem poses a significant challenge to the OptStrain methodology as the number of reaction database entries increase from a few to tens of thousands. At the expense of not enforcing stoichiometric balances, graph-based algorithms have inherently better-scaling properties for exhaustively identifying all min-path reaction entries that link a source with a target metabolite. Hatzimanikatis et. al. [72] introduced a graph-based heuristic approach (BNICE) to identify all possible biosynthetic routes from a given substrate to a target chemical by hypothesized enzymatic reaction rules. In addition, the BNICE framework was used to identify novel metabolic pathways for the synthesis of 3-hydroxypropionate in E. coli [73]. Based on a similar approach, a new scoring algorithm [74] was introduced to evaluate and compare novel pathways generated using enzyme-reaction rules. In addition, several techniques such as PathMiner [75], PathComp [76], Pathway Tools [77, 78], MetaRoute [79], PathFinder [80] and UM-BBD Pathway Prediction System [81] have been used to search databases for bioconversion routes.
We recently published [82] a graph-based algorithm that used reaction information from BRENDA and KEGG to exhaustively identify all connected paths from a source to a target metabolite using a customized min-path algorithm [83]. We first demonstrated the min-path procedure by identifying all synthesis routes for 1-butanol from pyruvate using a database of 9,921 reactions and 17,013 metabolites manually extracted from both BRENDA and KEGG. Here, we re-visited the same task using the full list of reactions and metabolites present in MetRxn to assess the discovery potential of using MetRxn. Figure 5 illustrates all identified pathways from pyruvate to 1-butanol before MetRxn (29, shown in blue) and the ones discovered after using MetRxn (112, shown in green). As many as 83 new avenues for 1-butanol production were revealed as a consequence of using the expanded and standardized MetRxn resource. In addition, the search algorithm recovered known [84–88] synthesis routes using E. coli for the production of 1-butanol (shown in orange). The first pathway involves the fermentative transformation of pyruvate and acetyl-CoA to 1-butanol using enzymes from C. acetobutylicum [89]. The second pathway uses ketoacid precursors [84]. This example demonstrates how the biotransformations stored in MetRxn can be used to traverse a multitude of production routes for targeted bioproducts.
Pathways from pyruvate to 1-butanol. Using the MetRxn knowledgebase, we identified a large number of new pathways (green) as well as previously established ones (orange) and those identified found in a previous study [82] (blue).
Conclusions
MetRxn enables the standardization, correction and utilization of rapidly growing metabolic information for over 76,000 metabolites participating in 72,000 reactions (including unresolved entries). The library of standardized and balanced reactions streamlines the process of reconstructing organism-specific metabolism and opens the way for identifying new paths for metabolic flux redirection. Moreover, the standardization of published genome-scale models enables the rapidly growing community of researchers who make use of metabolic information to understand metabolism at an organism-level and re-deploy it for various biotechnological objectives. By removing standardization and data heterogeneity bottlenecks the pace of knowledge creation and discovery from users of this resource will be accelerated. MetRxn is constructed in a way that allows for quick updating and tracking of changes that occur in the primary databases, as well as available parsing tools that allow for rapid import of new genome-scale metabolic models as they become available. By having exports in SBML, MetRxn's output can be directly interfaced with software packages such as the COBRA toolbox.
During the construction of the initial release of MetRxn, we managed to associate structures for over 8,800 metabolites and re-balanced more than 2,400 reaction instances across 44 metabolic models. This enables the genuine comparison of metabolic content between metabolic models. Preliminary results reinforce that that discrepancies between metabolic models echo not only genuine differences in metabolism but also assumptions and workflow followed by the model creator(s). Going forward, we will continue to expand MetRxn to include more genome-scale metabolic models and add additional tools to aid in their analysis. Because we anticipate that the scope and number of models will rapidly expand, we plan to invite and encourage the community to offer comments about metabolite and reaction information as well as provide feedback on MetRxn itself.
Availability and requirements
MetRxn is available at http://metrxn.che.psu.edu. Its use is freely available for all non-commercial activity.
References
Liolios K, Tavernarakis N, Hugenholtz P, Kyrpides NC: The Genomes On Line Database (GOLD) v.2: a monitor of genome projects worldwide. Nucleic Acids Res 2006, (34 database):D332–4.
Stolyar S, Van Dien S, Hillesland KL, Pinel N, Lie TJ, Leigh JA, Stahl DA: Metabolic modeling of a mutualistic microbial community. Mol Syst Biol 2007, 3: 92.
Reed JL, Famili I, Thiele I, Palsson BO: Towards multidimensional genome annotation. Nat Rev Genet 2006, 7(2):130–41. 10.1038/nrg1769
Burgard AP, Pharkya P, Maranas CD: Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng 2003, 84(6):647–57. 10.1002/bit.10803
Oliveira AP, Nielsen J, Forster J: Modeling Lactococcus lactis using a genome-scale flux model. BMC Microbiol 2005, 5: 39. 10.1186/1471-2180-5-39
Alper H, Jin YS, Moxley JF, Stephanopoulos G: Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab Eng 2005, 7(3):155–64. 10.1016/j.ymben.2004.12.003
Pharkya P, Maranas CD: An optimization framework for identifying reaction activation/inhibition or elimination candidates for overproduction in microbial systems. Metab Eng 2006, 8(1):1–13. 10.1016/j.ymben.2005.08.003
Almaas E, Kovacs B, Vicsek T, Oltvai ZN, Barabasi AL: Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature 2004, 427(6977):839–43. 10.1038/nature02289
Burgard AP, Nikolaev EV, Schilling CH, Maranas CD: Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res 2004, 14(2):301–12. 10.1101/gr.1926504
Motter AE, Gulbahce N, Almaas E, Barabasi AL: Predicting synthetic rescues in metabolic networks. Mol Syst Biol 2008, 4: 168.
Jin YS, Jeffries TW: Stoichiometric network constraints on xylose metabolism by recombinant Saccharomyces cerevisiae. Metab Eng 2004, 6(3):229–38. 10.1016/j.ymben.2003.11.006
Lee DY, Fan LT, Park S, Lee SY, Shafie S, Bertok B, Friedler F: Complementary identification of multiple flux distributions and multiple metabolic pathways. Metab Eng 2005, 7(3):182–200. 10.1016/j.ymben.2005.02.002
Jamshidi N, Palsson BO: Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst Biol 2007, 1: 26. 10.1186/1752-0509-1-26
Reed JL, Patel TR, Chen KH, Joyce AR, Applebee MK, Herring CD, Bui OT, Knight EM, Fong SS, Palsson BO: Systems approach to refining genome annotation. Proc Natl Acad Sci USA 2006, 103(46):17480–4. 10.1073/pnas.0603364103
Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Sohngen C, Stelzer M, Thiele J, Schomburg D: BRENDA, the enzyme information system in 2011. Nucleic Acids Res 2011, (39 database):D670–6.
Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M: KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 2009, (38 database):D355–60.
Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Paley S, Popescu L, Pujar A, Shearer AG, Zhang P, Karp PD: The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 2009, (38 database):D473–9.
Lang M, Stelzer M, Schomburg D: BKM-react, an integrated biochemical reaction database. BMC Biochem 2011, 12: 42. 10.1186/1471-2091-12-42
Gao J, Ellis LB, Wackett LP: The University of Minnesota Biocatalysis/Biodegradation Database: improving public access. Nucleic Acids Res 2010, (38 database):D488–91.
Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO: Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol 2009, 7(2):129–43.
Gevorgyan A, Poolman MG, Fell DA: Detection of stoichiometric inconsistencies in biomolecular models. Bioinformatics 2008, 24(19):2245–51. 10.1093/bioinformatics/btn425
Notebaart RA, van Enckevort FH, Francke C, Siezen RJ, Teusink B: Accelerating the reconstruction of genome-scale metabolic networks. BMC Bioinformatics 2006, 7: 296. 10.1186/1471-2105-7-296
Ott MA, Vriend G: Correcting ligands, metabolites, and pathways. BMC Bioinformatics 2006, 7: 517. 10.1186/1471-2105-7-517
Fleischmann A, Darsow M, Degtyarenko K, Fleischmann W, Boyce S, Axelsen KB, Bairoch A, Schomburg D, Tipton KF, Apweiler R: IntEnz, the integrated relational enzyme database. Nucleic Acids Res 2004, (32 database):D434–7.
Bairoch A: The ENZYME database in 2000. Nucleic Acids Res 2000, 28(1):304–5. 10.1093/nar/28.1.304
Vastrik I, D'Eustachio P, Schmidt E, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, Wu G, Birney E, Stein L: Reactome: a knowledge base of biologic pathways and processes. Genome Biol 2007, 8(3):R39. 10.1186/gb-2007-8-3-r39
Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, Kanapin A, Lewis S, Mahajan S, May B, Schmidt E, Vastrik I, Wu G, Birney E, Stein L, D'Eustachio P: Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res 2009, (37 database):D619–22.
Stobbe MD, Houten SM, Jansen GA, van Kampen AH, Moerland PD: Critical assessment of human metabolic pathway databases: a stepping stone for future integration. BMC Syst Biol 2011, 5: 165. 10.1186/1752-0509-5-165
Bornstein BJ, Keating SM, Jouraku A, Hucka M: LibSBML: an API library for SBML. Bioinformatics 2008, 24(6):880–1. 10.1093/bioinformatics/btn051
Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Le Novere N, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J: The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 2003, 19(4):524–31. 10.1093/bioinformatics/btg015
Stromback L, Lambrix P: Representations of molecular pathways: an evaluation of SBML, PSI MI and BioPAX. Bioinformatics 2005, 21(24):4401–7. 10.1093/bioinformatics/bti718
Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BO: A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol 2007, 3: 121.
Radrich K, Tsuruoka Y, Dobson P, Gevorgyan A, Swainston N, Baart G, Schwartz JM: Integration of metabolic databases for the reconstruction of genome-scale metabolic networks. BMC Syst Biol 2010, 4: 114. 10.1186/1752-0509-4-114
Pitkanen E, Akerlund A, Rantanen A, Jouhten P, Ukkonen E: ReMatch: a web-based tool to construct, store and share stoichiometric metabolic models with carbon maps for metabolic flux analysis. J Integr Bioinform 2008., 5(2):
Quek LE, Nielsen LK: On the reconstruction of the Mus musculus genome-scale metabolic network model. Genome Inform 2008, 21: 89–100.
Gonzalez O, Gronau S, Falb M, Pfeiffer F, Mendoza E, Zimmer R, Oesterhelt D: Reconstruction, modeling & analysis of Halobacterium salinarum R-1 metabolism. Mol Biosyst 2008, 4(2):148–59. 10.1039/b715203e
Henry CS, DeJongh M, Best AA, Frybarger PM, Linsay B, Stevens RL: High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol 2010, 28(9):977–82. 10.1038/nbt.1672
Weininger D: SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. Journal of Chemical Information and Computer Sciences 1988, 28(1):31–36.
Weininger D, Weininger A, Weininger JL: SMILES. 2. Algorithm for generation of unique SMILES notation. Journal of Chemical Information and Computer Sciences 1989, 29(2):97–101.
Daylight Theory Manual[http://www.daylight.com/dayhtml/doc/theory/]
Weininger D: SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. Journal of chemical information and computer sciences 1988, 28(1):31.
Weininger D, Weininger A, Weininger J: SMILES. 2. Algorithm for generation of unique SMILES notation. J Chem Inf Comput Sci 1989, 29: 97–101. 10.1021/ci00062a008
Varma A, Palsson BO: Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl Environ Microbiol 1994, 60(10):3724–31.
Kjeldsen KR, Nielsen J: In silico genome-scale reconstruction and validation of the Corynebacterium glutamicum metabolic network. Biotechnol Bioeng 2009, 102(2):583–97. 10.1002/bit.22067
Durot M, Le Fevre F, de Berardinis V, Kreimeyer A, Vallenet D, Combe C, Smidtas S, Salanoubat M, Weissenbach J, Schachter V: Iterative reconstruction of a global metabolic model of Acinetobacter baylyi ADP1 using high-throughput growth phenotype and gene essentiality data. BMC Syst Biol 2008, 2: 85. 10.1186/1752-0509-2-85
Henry CS, Zinner JF, Cohoon MP, Stevens RL: iBsu1103: a new genome-scale metabolic model of Bacillus subtilis based on SEED annotations. Genome Biol 2009, 10(6):R69. 10.1186/gb-2009-10-6-r69
David H, Ozcelik IS, Hofmann G, Nielsen J: Analysis of Aspergillus nidulans metabolism at the genome-scale. BMC Genomics 2008, 9: 163. 10.1186/1471-2164-9-163
Teusink B, Wiersma A, Molenaar D, Francke C, de Vos WM, Siezen RJ, Smid EJ: Analysis of growth of Lactobacillus plantarum WCFS1 on a complex medium using a genome-scale metabolic model. J Biol Chem 2006, 281(52):40041–8. 10.1074/jbc.M606263200
Oberhardt MA, Puchalka J, Fryer KE, Martins dos Santos VA, Papin JA: Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. J Bacteriol 2008, 190(8):2790–803. 10.1128/JB.01583-07
Becker SA, Palsson BO: Genome-scale reconstruction of the metabolic network in Staphylococcus aureus N315: an initial draft to the two-dimensional annotation. BMC Microbiol 2005, 5: 8. 10.1186/1471-2180-5-8
Oh YK, Palsson BO, Park SM, Schilling CH, Mahadevan R: Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J Biol Chem 2007, 282(39):28791–9. 10.1074/jbc.M703759200
Nogales J, Palsson BO, Thiele I: A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC Syst Biol 2008, 2: 79. 10.1186/1752-0509-2-79
JL Reed, Encarnacion S, Collado-Vides J, Palsson BO: Metabolic reconstruction and modeling of nitrogen fixation in Rhizobium etli. PLoS Comput Biol 2007, 3(10):1887–95.
Duarte NC, Herrgard MJ, Palsson BO: Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res 2004, 14(7):1298–309. 10.1101/gr.2250904
Herrgard MJ, Swainston N, Dobson P, Dunn WB, Arga KY, Arvas M, Bluthgen N, Borger S, Costenoble R, Heinemann M, Hucka M, Le Novere N, Li P, Liebermeister W, Mo ML, Oliveira AP, Petranovic D, Pettifer S, Simeonidis E, Smallbone K, Spasic I, Weichart D, Brent R, Broomhead DS, Westerhoff HV, Kirdar B, Penttila M, Klipp E, Palsson BO, Sauer U, Oliver SG, Mendes P, Nielsen J, Kell DB: A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol 2008, 26(10):1155–60. 10.1038/nbt1492
Reed JL, Vo TD, Schilling CH, Palsson BO: An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol 2003, 4(9):R54. 10.1186/gb-2003-4-9-r54
Feist AM, Scholten JC, Palsson BO, Brockman FJ, Ideker T: Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol Syst Biol 2006, 2: 2006 0004.
Edwards JS, Palsson BO: The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc Natl Acad Sci USA 2000, 97(10):5528–33. 10.1073/pnas.97.10.5528
Sheikh K, Forster J, Nielsen LK: Modeling hybridoma cell metabolism using a generic genome-scale metabolic model of Mus musculus. Biotechnol Prog 2005, 21(1):112–21.
Kuepfer L, Sauer U, Blank LM: Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res 2005, 15(10):1421–30. 10.1101/gr.3992505
Forster J, Famili I, Fu P, Palsson BO, Nielsen J: Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res 2003, 13(2):244–53. 10.1101/gr.234503
Kim TY, Kim HU, Park JM, Song H, Kim JS, Lee SY: Genome-scale analysis of Mannheimia succiniciproducens metabolism. Biotechnol Bioeng 2007, 97(4):657–71. 10.1002/bit.21433
Borodina I, Krabben P, Nielsen J: Genome-scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res 2005, 15(6):820–9. 10.1101/gr.3364705
Schilling CH, Covert MW, Famili I, Church GM, Edwards JS, Palsson BO: Genome-scale metabolic model of Helicobacter pylori 26695. J Bacteriol 2002, 184(16):4582–93. 10.1128/JB.184.16.4582-4593.2002
Lee J, Yun H, Feist AM, Palsson BO, Lee SY: Genome-scale reconstruction and in silico analysis of the Clostridium acetobutylicum ATCC 824 metabolic network. Appl Microbiol Biotechnol 2008, 80(5):849–62. 10.1007/s00253-008-1654-4
Roberts SB, Gowen CM, Brooks JP, Fong SS: Genome-scale metabolic analysis of Clostridium thermocellum for bioethanol production. BMC Syst Biol 2010, 4: 31. 10.1186/1752-0509-4-31
Bode HB, Muller R: The impact of bacterial genomics on natural product research. Angew Chem Int Ed Engl 2005, 44(42):6828–46. 10.1002/anie.200501080
Wenzel SC, Muller R: Recent developments towards the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr Opin Biotechnol 2005, 16(6):594–606. 10.1016/j.copbio.2005.10.001
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y: KEGG for linking genomes to life and the environment. Nucleic Acids Res 2008, (36 database):D480–4.
Chang A, Scheer M, Grote A, Schomburg I, Schomburg D: BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res 2009, (37 database):D588–92.
Pharkya P, Burgard AP, Maranas CD: OptStrain: a computational framework for redesign of microbial production systems. Genome Res 2004, 14(11):2367–76. 10.1101/gr.2872004
Hatzimanikatis V, Li C, Ionita JA, Henry CS, Jankowski MD, Broadbelt LJ: Exploring the diversity of complex metabolic networks. Bioinformatics 2005, 21(8):1603–9. 10.1093/bioinformatics/bti213
Henry CS, Broadbelt LJ, Hatzimanikatis V: Discovery and analysis of novel metabolic pathways for the biosynthesis of industrial chemicals: 3-hydroxypropanoate. Biotechnol Bioeng 2010, 106(3):462–73.
Cho A, Yun H, Park JH, Lee SY, Park S: Prediction of novel synthetic pathways for the production of desired chemicals. BMC Systems Biology 2010., 4(35):
S Rao, Shah I: PathMiner: predicting metabolic pathways by heuristic search. Bioinformatics 2003, 19(13):1692–8. 10.1093/bioinformatics/btg217
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M: From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 2006, (34 database):D354–7.
Karp PD, Paley S, Romero P: The Pathway Tools software. Bioinformatics 2002, 18(Suppl 1):S225–32. 10.1093/bioinformatics/18.suppl_1.S225
Karp PD, Paley SM, Krummenacker M, Latendresse M, Dale JM, Lee TJ, Kaipa P, Gilham F, Spaulding A, Popescu L, Altman T, Paulsen I, Keseler IM, Caspi R: Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinform 2009.
Blum T, Kohlbacher O: MetaRoute: fast search for relevant metabolic routes for interactive network navigation and visualization. Bioinformatics 2008, 24(18):2108–9. 10.1093/bioinformatics/btn360
Goesmann A, Haubrock M, Meyer F, Kalinowski J, Giegerich R: PathFinder: reconstruction and dynamic visualization of metabolic pathways. Bioinformatics 2002, 18(1):124–9. 10.1093/bioinformatics/18.1.124
Ellis LB, Roe D, Wackett LP: The University of Minnesota Biocatalysis/Biodegradation Database: the first decade. Nucleic Acids Res 2006, (34 database):D517–21.
Ranganathan S, Maranas CD: Microbial 1-butanol production: Identification of non-native production routes and in silico engineering interventions. Biotechnol J 2010, 5(7):716–25. 10.1002/biot.201000171
Yen JY: Finding K Shortest Loopless Paths in a Network. Management Science Series a-Theory 1971, 17(11):712–716.
Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJY, Hanai T, Liao JC: Metabolic engineering of Escherichia coli for 1-butanol production. Metabolic Engineering 2008, 10(6):305–311. 10.1016/j.ymben.2007.08.003
Formanek J, Mackie R, Blaschek HP: Enhanced Butanol Production by Clostridium beijerinckii BA101 Grown in Semidefined P2 Medium Containing 6 Percent Maltodextrin or Glucose. Appl Environ Microbiol 1997, 63(6):2306–10.
Lee JY, Jang YS, Lee J, Papoutsakis ET, Lee SY: Metabolic engineering of Clostridium acetobutylicum M5 for highly selective butanol production. Biotechnol J 2009, 4(10):1432–40. 10.1002/biot.200900142
Shen CR, Liao JC: Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab Eng 2008, 10(6):312–20. 10.1016/j.ymben.2008.08.001
Sillers R, Chow A, Tracy B, Papoutsakis ET: Metabolic engineering of the non-sporulating, non-solventogenic Clostridium acetobutylicum strain M5 to produce butanol without acetone demonstrate the robustness of the acid-formation pathways and the importance of the electron balance. Metab Eng 2008, 10(6):321–32. 10.1016/j.ymben.2008.07.005
Atsumi S, Hanai T, Liao JC: Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 2008, 451(7174):86–9. 10.1038/nature06450
Acknowledgements and Funding
This work was funded by DOE grant DE-FG02-05ER25684. The authors would like to gratefully acknowledge Robert Pantazes, Sridhar Ranganatha, Rajib Saha, and Alireza Zomorrodi for their help with testing and feedback on the MetRxn web interface.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
AK generated the software and tools for MetRxn and assisted in drafting the manuscript. PFS participated in the design of the database, performed database curation and FBA analysis, and drafted the manuscript. CDM conceived the study, participated in the design of the database and edited the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kumar, A., Suthers, P.F. & Maranas, C.D. MetRxn: a knowledgebase of metabolites and reactions spanning metabolic models and databases. BMC Bioinformatics 13, 6 (2012). https://doi.org/10.1186/1471-2105-13-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2105-13-6