Abstract
RNA synthesis and decay counteract each other and therefore inversely regulate gene expression in pro- and eukaryotic cells by controlling the steady-state level of individual transcripts. Genetic and biochemical data together with recent in depth annotation of bacterial genomes indicate that many components of the bacterial RNA decay machinery are evolutionarily conserved and that their functional analogues exist in organisms belonging to all kingdoms of life. Here we briefly review biological functions of essential enzymes, their evolutionary conservation and multienzyme complexes that are involved in mRNA decay in Escherichia coli and discuss their conservation in evolutionarily distant bacteria.
Similar content being viewed by others
1. mRNA turnover and its role in gene expression
In contrast to metabolically stable DNA serving as a storehouse of genetic information, the fraction of total RNA that delivers coding information to the protein-synthesizing machinery (i.e. mRNA) is intrinsically labile and continuously synthesized. The steady-state level of mRNA is tightly controlled enabling bacteria to selectively copy (transcribe) and decode genetic information pertinent to a particular physiological state (Figure 1). Since the steady-state level of mRNA can vary and is a function of RNA synthesis and decay, the control of mRNA stability plays an essential role in the regulation of gene expression. As transcription and translation are coupled in bacteria, the degree of their coupling can control the access of individual transcripts to the RNA decay machinery, thus influencing the rate of mRNA turnover. For more information about the crosstalk between translation and mRNA decay in bacteria and its regulation by environmental factors, we recommend some recent reviews (see [1–5]).
RNA synthesis and turnover as part of the gene expression network in bacteria. Different types of RNA (mRNAs, ribosomal and transfer RNA pre-cursors and various non-coding RNAs) either can directly be involved in translation (e.g. mRNAs) or undergo further processing (pre-cursors of stable RNA) or degradation (untranslated or poorly translated mRNAs) by the RNA decay machinery. The final products of RNA turnover, mononucleotides, are used for the next cycles of RNA synthesis (recycling).
The ability of bacteria to rely on remarkably diverse metabolic pathways in order to adopt and strive in different environmental niches suggests that the nature and number of enzymatic activities involved in specific metabolic pathways including mRNA turnover can greatly vary from species to species. Hence, an analysis of the putative organization and composition of bacterial mRNA decay machineries that belong to phylogenetically distant species should enable us to gain critical insights into the evolution of RNA decay pathways and their conservation in bacteria. The main objective of this review was therefore to assess the evolutionary conservation of RNases and ancillary factors that are involved in mRNA turnover and briefly discuss their specific roles in this process.
2. Enzymes with major and ancillary functions in mRNA turnover and their phylogenetic conservation in bacteria
Early studies on RNA processing and decay in E. coli, a Gram-negative bacterium that belongs to the gamma division of proteobacteria, revealed several endoribonucleases (cleave RNA internally), exoribonucleases (sequentially remove mononucleotides from either the 5' or the 3'-end of RNA) and other RNA-modifying enzymes with important functions in mRNA turnover (Table 1). The specific roles of these enzymes as well as their functional homologues found in another model organism, the Gram-positive bacterium Bacillus subtilis, have been reviewed recently [5]. Here, we focus on the phylogenetic conservation of the major RNases (e.g., RNase E, polynucleotide phosphorylase, RNase II) and ancillary RNA-modifying enzymes (RNA pyrophosphohydrolase (RppH), poly(A) polymerase I (PAPI) and RNA helicase B (RhlB)) involved in the turnover of mRNAs in bacteria. Previous bioinformatic approaches have revealed that several mRNA-degrading enzymes are not strictly conserved and can be absent in some classes of bacteria [6, 7]. The availability of new genomic data and discovery of novel RNases in bacteria prompted us to re-assess the phylogenetic conservation of these enzymes in bacterial species for which the sequence of the entire genome is available. The potential presence of mRNA degrading and mRNA-modifying ancillary enzymes was examined in all classes of bacteria by searching for the corresponding annotated genes and protein sequences available in the NCBI database http://www.ncbi.nlm.nih.gov/. The result of this analysis leads to several important conclusions regarding the nature and occurrence of RNases, ancillary enzymes (see 2.1 and 2.2) and their multienzyme assemblies (see 2.3) in evolutionarily distant species.
2.1 Conservation and diversity of major enzymes controlling the endoribonucleolytic decay of mRNA
Despite their indispensable functions in the processing of ribosomal and transfer RNA in E. coli, three major endoribonucleases, RNase E, RNase III and RNase P unequally contribute to mRNA decay. With few exceptions [8, 9], the endoribonucleolytic decay of E. coli transcripts primarily involves RNase E and sometimes RNase III (reviewed in [10]). Moreover, previous studies of mRNA decay pathways in E. coli demonstrated the key role of RNase E, a member of the RNase E/G family of ribonucleases, in carrying out the first endoribonucleolytic cleavages initiating the ribonucleolytic decay of E. coli transcripts (reviewed in [11]). Although homologues of RNase E/G are predicted to be present in many bacterial species, they are either partially or completely absent in some phyla of bacteria (Figure 2). The lack of genes coding for this endoribonuclease suggests that either (i) the main functions of RNase E/G are occasionally taken over by other endoribonucleases or that (ii) RNase E/G is redundant for RNA processing and decay in some species.
The phylogenetic distribution of main ribonucleases (RNase E/G, RNase III, RNases J1/J2, RNase Y, RNase PH, PNPase, RNase R, RNase II, Oligoribonuclease) and ancillary RNA modifying enzymes (RppH, PAPI, DEAD-box helicases) involved in the disintegration and turnover of bacterial transcripts are indicated by colored filled circles (from 'a' to 'l', respectively). The percentage of organisms in each phylum/class of bacteria for which the presence of each particular enzyme has been predicted by searching the NCBI database is indicated by differentially colored circles. The data are compiled based on analysis of completely sequenced genomes (1217 complete genome sequences available by 4 November 2010). Draft assemblies of genomes and hypothetical proteins were excluded from the analysis.
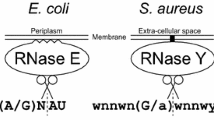
The first possibility is supported by a recent analysis of RNA processing and decays in B. subtilis (class Firmicutes) [12–14]. Despite the discovery of RNase E-like cleavages in this bacterium [15], they were subsequently attributed to the action of two B. subtilis endoribonucleases (RNases J1 and J2) that bear primarily functional rather than sequence homology to their E. coli counterpart. Both RNase J1 and J2 were suggested to functionally represent RNase E/G in B. subtilis by mimicking the ability of RNase E to make endoribonucleolytic cuts in a 5'-end-dependent manner [12] as well as its property to form multienzyme complexes [13, 14]. Interestingly, one recent study reported the existence and characterization of another B. subtilis endoribonuclease, RNase Y, and suggested that this enzyme is also functionally related to RNase E/G, in particular with regard to its role in mRNA turnover [16]. Consistent with this suggestion, we found that RNase Y appears to occur more frequently than RNases J1/J2 in the phyla that lack RNase E/G (Figure 2).
In contrast to Firmicutes, Actinomycetes and other phylas of bacteria whose members can apparently survive without RNase E/G by using its functional homologues, RNase Y and/or RNases J1/J2, some bacterial species seem to be able to carry out RNA processing and decay even in the absence of all these endoribonuclases (i.e., RNase E/G, RNase Y, and RNases J1/J2). Examples are some pathogenic bacteria that belong to the clades of Deinococcus, Dictyoglomy, Spirochaetales and Tenericutes. Many of these pathogens lack genes encoding not only the above endoribonucleases but also many exonucleases (see also 2.2).
Several studies revealed that the 5'-phosphorylation status of mRNA can control the efficiency of cleavages by RNase E/G homologues [17–21] as well as by RNases J1/J2 [12] and RNase Y [16]. As the E. coli pyrophosphohydrolase RppH (initially designated NudH/YgdP) is able to facilitate RNase E cleavage of primary transcripts by 5' pyrophosphate removal [22], we examined the presence of nudH/ygdP genes in genomes of phylogenetically distant bacteria. Despite the apparent absence of these genes in many classes of bacteria (Figure 2), their homologues that belong to the same family of Nudix hydrolases are known to be widely present in all three domains of life (reviewed in [23]). Therefore, it seems likely that the RNA pyrophosphohydrolase-mediated stimulation of mRNA decay in some bacterial species involves other members of the Nudix family of hydrolases.
2.2 Conservation and diversity of major enzymes controlling exoribonucleolytic decay of mRNA
A search for putative homologues of the three major mRNA-degrading exoribonucleases of E. coli (polynucleotide phosphorylase (PNPase), RNase II and RNase R) in other bacteria revealed that the corresponding genes can be found in nearly every class of bacteria (Figure 2). Although these observations suggest that mRNA decay in the majority of bacteria could be dependent on all three exoribonucleases, the actual contribution of each exoribonuclease to mRNA decay in these species may differ, as anticipated from previous studies of exonucleolytic decay of mRNA in B. subtilis (Firmicutes) and E. coli (Proteobacteria). These studies revealed that, in contrast to apparently similar roles of RNase II and PNPase in the degradation of E. coli mRNA [24], only PNPase plays a central role in the 3'-exonucleolytic decay of B. subtilis mRNA [25] with apparently less significant contribution of other exoribonucleases [25] including RNase PH [26], RNase R [27] and YhaM [28]. This is consistent with the previous finding that the 3'-to-5' exonuleolytic mRNA decay in B. subtilis, contrary to RNA turnover in E. coli, primarily proceeds through an "energy-saving" phosphorolytic pathway [29] mediated by PNPase. Further studies will be necessary to address systematically how phylogenetically distant bacteria combine different sets of exoribonucleases to carry out mRNA decay. Finally, given the high degree of phylogenetic conservation of PNPase and RNase II, it seems reasonable that one of the key ancillary enzymes, PAPI, which assists PNPase and RNase II in the degradation of structured RNAs, is likewise present in most of the bacteria, as shown in Figure 2.
2.3. Conservation of mRNA-degrading multienzyme complexes
Many E. coli mRNAs have relatively short half-lives (2-4 min) and are normally degraded in vivo without accumulation of intermediate products (reviewed in [30]), a phenomenon frequently referred to as the 'all-or-nothing' mechanism of mRNA turnover. The high processivity of mRNA decay is often discussed with reference to the coordinated action of ribonucleolytic enzymes and ancillary proteins that can associate with each other to form multienzyme ribonucleolytic complexes such as the E. coli degradosome (Figure 3A, [31–33]) and the bacterial exosome-like complex (Figure 3B) [34, 35]. Analyses of the E. coli degradosome revealed that RNase E serves as a "scaffolding" protein, through the C-terminal part of which other interacting protein partners such as PNPase (exoribonuclease), RhlB (DEAD-box helicase) and enolase (glycolytic enzyme) are bound [36, 37]. Consistent with these reports, the existence of functional interactions between the major components of the degradosome was confirmed in vivo[38–43] and in vitro[33, 44]. Apart from binding to RNase E, two major components of the E. coli degradosome, PNPase and RhlB helicase, were shown to form a complex resembling the eukaryotic exosome, a multienzyme assembly with RNA-hydrolyzing and RNA-unwinding activities (reviewed in [35]). The formation and functions of this complex in E. coli may not be unusual as both enzymes appear to exist in excess to RNase E in vivo and therefore can be involved in alternative protein-protein interactions. However, the actual contribution of this complex to RNA metabolism in bacteria remains to be determined. mRNA molecules that are degraded by these multiprotein assemblies (i.e., degradosome and exosome) are simultaneously exposed to several ribonucleolytic and other RNA-modifying activities and therefore undergo fast and coordinated decay without accumulation of detectable amounts of intermediate products.
Bacterial mRNA decay machineries. (A) The RNA degradosome is a multicomponent ribonucleolytic complex that includes an endoribonuclease (RNase E), a 3'→5' exoribonuclease (polynucleotide phosphorylase (PNPase)), a DEAD-box RNA helicase (RhlB helicase), and the glycolytic enzyme enolase [31–33]). (B) In E. coli, PNPase is associated with the RhlB independently of the RNA degradosome to form an evolutionarily conserved RNA-degradation machine termed as the "bacterial exosome" [34, 35]. This complex was shown to catalyze the 3'→ 5' exonucleolytic degradation of RNA using RhlB as cofactor to unwind structured RNA in an ATP-dependent manner.
Although significant progress has been achieved in the characterization of the E. coli degradosome (reviewed in [45]), our current knowledge of the composition and properties of similar complexes in other bacteria is still very limited. A previous comparison of RNase E/G sequences revealed that the C-terminal half of E. coli RNase E (residues 499-1061), which is involved in protein-protein interactions with other major components of the E. coli degradosome, is poorly conserved among RNase E/G homologues [36]. Despite the overall lack of conservation, the PNPase-binding site of E. coli RNase E (residues 1021-1061, see [37]) is known to possess high similarity to a short amino acid sequence found in H. influenza Rd RNase E (residues 896-927, [36]). Moreover, this sequence is highly conserved among RNase E/G homologues of certain γ-proteobacteria (e.g., Erwinia, Shigella, and Citrobacter) and therefore is presently annotated in the NCBI database as the PNPase-binding domain. The conservation of this domain (although primarily in enterobacterial species) is also supported by a recent analysis of Vibrio angustum S14 RNase E [46]. This study defined the last 80 amino acids at the C-terminus of Vibrio angustum S14 RNase E as the potential site for PNPase binding and revealed the putative enolase-binding domain, a region also highly conserved amongst enterobacteria [47, 48]. Collectively, the above findings and genomic data suggest that degradosome-like complexes are widespread in enterobacteria and organized in a similar manner.
In contrast to the apparently similar organization of enterobacterial degradosomes, their counterparts in other subclasses of γ-proteobacteria are less conserved. For instance, an analysis of the degradosome composition in the psychrotolerant γ-proteobacterium Pseudoalteromonas haloplanktis revealed that RNase E associates with PNPase and RhlB but not with enolase [49]. Moreover, a different degradosome-like complex consisting of RNase E, the hydrolytic exoribonuclease RNase R, and the DEAD-box helicase RhlE was purified from another psychrotrophic γ-proteobacterium, Pseudomonas syringae Lz4W [50]. As RNA structures are more stable at low temperatures and RNase R can degrade structural RNAs more efficiently than PNPase [51], the presence of RNase R (rather than PNPase) in this complex may be more advantageous for the degradosome-mediated decay in this psychrotrophic bacterium. RNase E-based degradosomes have also been isolated from other subclasses of proteobacteria. Hardwick and co-workers have recently isolated and characterized an RNase E-containing complex from the Gram-negative α-proteobacterium Caulobacter crescentus[52]. Apart from RNase E, this complex was found to contain PNPase, a DEAD-box RNA helicase and aconitase, an iron-dependent enzyme involved in the tricarboxylic acid cycle. One can envisage that, similar to its mycobacterial counterpart [53], C. crescentus aconitase may possess RNA-binding properties, and therefore can potentially modulate the efficiency and/or specificity of the degradosome-mediated RNA decay. More significant differences in the composition of degradosomes can be found in other α-proteobacteria. It has been shown that RNase E of Rhodobacter capsulatis forms a degradosome-like complex with two DEAD-box RNA helicases of 74 and 65 kDa and the transcription termination factor Rho [54]. Thus, the degradosome-dependent mRNA decay appears to involve different combinations of enzymatic activities even within the same class of bacteria.
In addition to analyzing the composition of degradosome complexes in Proteobacteria, some efforts were dedicated to identify degradosome-like complexes in Actinobacteria. These studies revealed that, similar to their E. coli counterpart, RNase E/G homologues can interact with PNPase in Streptomyces[55] and are able to co-purify with GroEL and metabolic enzymes in Mycobacteria[56]. The specific role of these polypeptides in RNA metabolism and the degree, to which their interaction with RNase E/G is conserved in Actinobacteria, remains to be established.
Aside from degradosome complexes that are believed to function in Proteobacteria and Actinobacteria, the existence of RNase E-based degradosomes in other classes of bacteria remains questionable. The small size (ca. 450-600 a.a., see Table 2) of RNase E/G homologues in many other classes of bacteria indicate that they primarily contain the evolutionarily conserved catalytic core of the enzyme and appear to lack regions serving as scaffolds for degradosome assembly [36, 57].
Interestingly, recent studies demonstrated that the Gram-positive bacterium B. subtilis (Firmicutes) possesses degradosome-like complexes, in which RNase E is represented by its functional homologues, RNases J1/J2 and RNase Y, interacting with PNPase, phosphofructokinase and enolase [13]. Further characterization of these complexes and elucidation of their specific roles in mRNA decay in B. subtilis and related species can offer many important insights into the mechanisms underlying mRNA decay in Firmicutes, the largest group of Gram-positive bacteria that have been studied so far [58].
3. Current unified model for mRNA decay pathways in E. coli
3.1 Both endo- and exoribonucleases act cooperatively to control mRNA decay
Despite phylogenetic conservation (Figure 2) and their apparent diversity (for a review, see [10]), mRNA decay pathways in E. coli are believed to include a number of common enzymatic steps catalyzed by ribonucleases and several ancillary mRNA-modifying enzymes. To discuss the role of each enzyme, we will refer to a unified model of mRNA turnover. According to this model (Figure 4A), conversion of E. coli mRNAs into their primary decay intermediates is frequently initiated by endoribonucleolytic cuts catalyzed by endoribonucleases specific for single- (e.g., RNase E/G) or double-stranded (RNase III) RNA. This step can be preceded (but not always, see [59]) by pyrophosphate removal (see below). During the initial endoribonucleolytic step, bacterial RNase E/G (or its functional homologues, RNases J1/J2 or RNase Y) attacks the full-length monophosphorylated (or sometimes triphosphorylated [59]) mRNAs to generate primary decay intermediates that are further degraded cooperatively by the combined action of endo- and exoribonucleases (Figure 4A). In E coli, the later steps of mRNA decay were shown to involve PNPase and RNase II, or occasionally RNase R [51, 60], which further degrade mRNA decay intermediates to yield short oligonucleotides that are, in turn, converted to mononucleotides by oligoribonuclease [61].
Current unified model of mRNA decay pathways in Escherichia coli. (A) Schematic representation of major enzymatic steps involved in the disintegration and complete turnover of primary transcripts in E. coli. The decay of a regular transcript is usually initiated by endonucleolytic cleavage to generate primary decay intermediates that are further converted to short oligoribonucleotides by the combined action of exo- and endoribonucleases. The oligoribonucleotides are further degraded into mononucleotides by oligoribonuclease. (B) Ancillary enzymes facilitating mRNA turnover and their modes of action. Degradation of mRNA can be stimulated via pyrophosphate removal by RppH, which converts 5'-triphosporylated primary transcripts into their monophosphorylated variants, thus facilitating their endoribonucleolytic cleavage by RNase E [22, 76] or by RNases J1/J2 [12] or by RNase Y [16] in B. subtilis. As the action of exoribonucleases can be inhibited by 3'-terminal stem-loop structures, two groups of ancillary RNA-modifying enzymes, PAPI and RhlB, help exonucleases to overcome this inhibitory effect. PAPI exerts its action by adding short stretches of adenosine residues, thereby facilitating exonuclease binding and subsequent cleavage of structured RNAs [10]. Enzymes of the second group, DEAD-box helicases such as E. coli RhlB, increase the efficiency of the exonuclease-dependent decay by unwinding double-stranded RNA regions in an ATP-dependent fashion.
3.2 Ancillary enzymes facilitate mRNA turnover by assisting ribonucleases
In addition to the major degrading enzymes, a number of ancillary mRNA-modifying enzymes can facilitate mRNA turnover (Table 1). In fact, pyrophosphate removal at the 5'-end and addition of a single-stranded, poly(A) extension at the 3'-end are two critical steps in the mRNA decay pathway promoting mRNA cleavage in E. coli and presumably in other proteobacteria. In general, however, the participation of these enzymes in mRNA decay in some bacterial species or organelles is not required (see section 2). One of these enzymes, RppH, was shown to accelerate mRNA decay by converting the 5'-triphosphate group of primary transcripts to 5' monophosphate, thereby rendering mRNA species that are more efficiently recognized and cleaved by RNase E [17, 18] and RNase G [19].
Unlike RppH, whose action promotes endoribonucleolytic cleavages, some mRNA-modifying enzymes can stimulate degradation by 3' to 5' exonucleases (reviewed in [62, 63]). Previous work has shown that the 3' to 5' degradation of transcripts by PNPase and RNase II in E. coli proceeds only efficiently on unstructured mRNAs and is impeded by stable stem-loop structures occurring internally (e.g., in intergenic regions of polycistronic transcripts such as REP stabilizers found in the malEFG and many other intergenic regions [39]) or at the 3' end of bacterial transcripts (i.e., transcription terminators [64]). These structures typically cause exoribonuclease stalling and subsequent dissociation of exoribonucleases from decay intermediates (reviewed in [62, 63]). To prevent accumulation of decay intermediates that are resistant to 3' to 5' degradation by exoribonucleses, E. coli and apparently other bacteria employ a mechanism that increases the susceptibility of an mRNA decay intermediate to exonucleases by adding a poly(A) tail to its 3' end (Figure 4B). Consequently, repetitive cycles of poly(A) addition carried out by PAPI combined with exonuclease-catalyzed trimming was shown to result in the complete digestion of structured RNAs by either PNPase or RNase II in vitro[65]. Consistent with these findings, mRNA decay in a mutant lacking functional PAPI results in the accumulation of intermediate products of mRNA decay [64, 66–69], thus indicating that the addition of poly(A) tails is indeed required for the normal mRNA turnover in E. coli. Because several aspects of poly(A)-assisted mRNA turnover including its role in the decay of stable RNA fall beyond the scope of this review, the interested reader is referred to other work covering this topic [70].
In E. coli, the exonucleolytic decay of highly structured RNAs can also be assisted by the RhlB (Figure 4B). This enzyme unwinds RNA structures in an ATP-dependent manner and therefore facilitates their degradation by exonucleases in vivo[39] and in vitro[33, 34]. Moreover, RhlB is an integral part of the multienzyme RNA degradosome and exosome-like complexes and believed to exert its functions primarily as component of the mRNA decay machinery.
4. Conclusion and perspectives
A previous analysis of RNA processing/decay pathways in several distantly-related bacterial species including the two major model organisms, E. coli (Proteobacteria) and B. subtilis (Firmicutes) has identified the key ribonucleases involved in mRNA turnover in bacteria (reviewed in [5]). Herein, a search for their homologues in bacteria with completely sequenced genomes revealed that many components of the bacterial mRNA decay machinery (RNase III and three major exoribonucleases, PNPase, RNase II and RNase R) as well as PAPI and RhlB) are highly conserved across the bacterial kingdom (see Figure 2). In contrast, the major endoribonucleases RNase E/G, RNases J1/J2, and RNase Y possess only functional (but not sequence) conservation. Although they were found only in particular classes of bacteria, at least one of them is present in nearly every species. Thus, although RNA processing/decay in phylogenetically distant bacterial species is not necessarily carried out by the same set of ribonucleolytic enzymes (see previous sections), the minimal set of enzymatic activities (at least one functional homologue of RNase E/G and one 3' to 5' exoribonuclease) required for mRNA turnover in prokaryotic organisms is likely conserved in a vast majority of bacterial species.
Surprisingly, the number of enzymes with potential roles in RNA processing and decay is dramatically reduced in several intracellular pathogens possessing relatively small (less than 1 Mbp) genomes (e.g., Mycoplasma (Tenericutes), Rickettsia (α-Proteobacteria) and Chlamydia (Chlamydiae/Verrucomicrobia group)). In contrast to the presence of seven distinct exoribonucleases in E. coli, only one of them can be found in Mycoplasma (subclass Mollicutes (Tenericutes)). Analysis of RNA metabolism in Mycoplasma genitalium suggests that exonucleolytic decay in this bacterium can be accomplished by a single exoribonuclease, RNase R [71]. Another prominent feature of Mycoplasma is the lack of genomic sequences potentially encoding a homologues of E. coli PAPI known to catalyze the addition of poly(A) to the 3' end of E. coli transcripts [72]. The lack of this enzyme is consistent with the recent finding that demonstrated the absence of polyadenylated RNA in Mycoplasma [73]. Although the poly(A)-dependent enhancement of mRNA decay is likely redundant for some intracellular pathogens, it seems to be more important in some Proteobacteria and Firmicutes, as it can offer an additional mean to control the efficiency of mRNA turnover. In other words, unlike pathogens that continuously reside in host cells, bacteria that strive in highly diverse and continuously changing environments (e.g., Escherichia coli) use a large number of ribonucleases and ancillary mRNA-modifying enzymes such as poly(A) polymerases to efficiently regulate mRNA stability in response to environmental signals. Future studies addressing the main differences between the mechanisms of mRNA decay of intracellular pathogens and the currently used model organisms (E. coli and B. subtilis) may lead to important insights concerning the evolution of the mRNA decay machinery in bacteria.
Similar to other essential cellular processes controlling inheritance and expression of genetic information (i.e., DNA replication, transcription and translation), mRNA decay was found to be carried out by multienzyme complexes, several of which have been isolated from Proteobacteria, Actinobacteria and Firmicutes over the last two decades. The existence of functional interactions between the major components of the E. coli degradosome and their impact on mRNA turnover [38–43] suggest that multienzyme complexes (instead of a pull of non-interacting enzymes) are favorable for attaining a higher efficiency of mRNA decay. The role of similar complexes in other bacteria is still poorly defined. Moreover, we do not know to which degree RNase E/G-based degradosomes resemble their counterparts containing RNases J1/J2 or RNase Y existing in many other classes of bacteria. Likewise, the mechanisms modulating the composition, activity and specificity of these multienzyme assemblies in response to changing physiological conditions remain largely unknown and merit further analysis. Finally, although the last step of mRNA decay in E. coli has been shown to be accomplished by oligoribonuclease encoded by the orn gene [61], this gene is apparently absent in many other bacterial species (Figure 2). A search for activities that can degrade RNA oligoribonucleotides in Firmicutes lacking sequence homologues of E. coli oligoribonuclease led to the discovery of B. subtilis Ytq1 [74]. This enzyme possesses an oligoribonuclease-like activity and is able to complement the E. coli orn mutant; homologues of its gene are present in many bacteria [75]. Although Ytq1 can be considered as a functional homologue of oligoribonuclease, further efforts are needed to disclose the nature and distribution of functional homologues that may exist in bacterial species lacking both oligoribonuclease and Ytq1.
References
Deana A, Belasco JG: Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005, 19 (21): 2526-2533. 10.1101/gad.1348805.
Kaberdin VR, Bläsi U: Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol Reviews. 2006, 30: 967-979. 10.1111/j.1574-6976.2006.00043.x.
Richards J, Sundermeier T, Svetlanov A, Karzai AW: Quality control of bacterial mRNA decoding and decay. Biochim Biophys Acta. 2008, 1779 (9): 574-582.
Dreyfus M: Killer and protective ribosomes. Prog Mol Biol Transl Sci. 2009, 85: 423-466. full_text.
Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M: The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev. 2010, 34 (5): 883-923.
Condon C, Putzer H: The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 2002, 30 (24): 5339-5346. 10.1093/nar/gkf691.
Zuo Y, Deutscher MP: Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001, 29 (5): 1017-1026. 10.1093/nar/29.5.1017.
Alifano P, Rivellini F, Piscitelli C, Arraiano CM, Bruni CB, Carlomagno MS: Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic messenger RNA. Genes Dev. 1994, 8 (24): 3021-3031. 10.1101/gad.8.24.3021.
Li Y, Altman S: A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc Natl Acad Sci USA. 2003, 100 (23): 13213-13218. 10.1073/pnas.2235589100.
Coburn GA, Mackie GA: Degradation of mRNA in Escherichia coli: An old problem with some new twists. Prog Nucleic Acids Res Mol Biol. 1999, 62: 55-108. 10.1016/S0079-6603(08)60505-X.
Carpousis AJ, Luisi BF, McDowall KJ: Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Mol Biol Transl Sci. 2009, 85: 91-135. full_text.
Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H: Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005, 33 (7): 2141-2152. 10.1093/nar/gki505.
Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, Hammer E, Volker U, Stulke J: Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics. 2009, 8 (6): 1350-1360. 10.1074/mcp.M800546-MCP200.
Mathy N, Hebert A, Mervelet P, Benard L, Dorleans A, de la Sierra-Gallay IL, Noirot P, Putzer H, Condon C: Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol Microbiol. 2010, 75 (2): 489-498. 10.1111/j.1365-2958.2009.07004.x.
Condon C, Putzer H, Luo D, Grunberg-Manago M: Processing of the Bacillus subtilis thrS leader mRNA is RNase E-dependent in Escherichia coli. J Mol Biol. 1997, 268 (2): 235-242. 10.1006/jmbi.1997.0971.
Shahbabian K, Jamalli A, Zig L, Putzer H: RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 2009, 28 (22): 3523-3533. 10.1038/emboj.2009.283.
Mackie GA: Ribonuclease E is a 5'-end-dependent endonuclease. Nature. 1998, 395 (6703): 720-723. 10.1038/27246.
Mackie GA: Stabilization of circular rpsT mRNA demonstrates the 5'-end dependence of RNase E action in vivo. J Biol Chem. 2000, 275 (33): 25069-25072. 10.1074/jbc.C000363200.
Jourdan SS, McDowall KJ: Sensing of 5' monophosphate by Escherichia coli RNase G can significantly enhance association with RNA and stimulate the decay of functional mRNA transcripts in vivo. Mol Microbiol. 2008, 67 (1): 102-115. 10.1111/j.1365-2958.2007.06028.x.
Kaberdin VR, Bizebard T: Characterization of Aquifex aeolicus RNase E/G. Biochem Biophys Res Commun. 2005, 327 (2): 382-392. 10.1016/j.bbrc.2004.12.017.
Zeller ME, Csanadi A, Miczak A, Rose T, Bizebard T, Kaberdin V: Quaternary structure and biochemical properties of mycobacterial RNase E/G. Biochem J. 2007, 403: 207-215. 10.1042/BJ20061530.
Deana A, Celesnik H, Belasco JG: The bacterial enzyme RppH triggers messenger RNA degradation by 5' pyrophosphate removal. Nature. 2008, 451 (7176): 355-358. 10.1038/nature06475.
McLennan AG: The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006, 63 (2): 123-143. 10.1007/s00018-005-5386-7.
Donovan WP, Kushner SR: Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci USA. 1986, 83 (1): 120-124. 10.1073/pnas.83.1.120.
Oussenko IA, Abe T, Ujiie H, Muto A, Bechhofer DH: Participation of 3'-to-5' exoribonucleases in the turnover of Bacillus subtilis mRNA. J Bacteriol. 2005, 187 (8): 2758-2767. 10.1128/JB.187.8.2758-2767.2005.
Craven MG, Henner DJ, Alessi D, Schauer AT, Ost KA, Deutscher MP, Friedman DI: Identification of the rph (RNase PH) gene of Bacillus subtilis: evidence for suppression of cold-sensitive mutations in Escherichia coli. J Bacteriol. 1992, 174 (14): 4727-4735.
Oussenko IA, Bechhofer DH: The yvaJ gene of Bacillus subtilis encodes a 3'-to-5' exoribonuclease and is not essential in a strain lacking polynucleotide phosphorylase. J Bacteriol. 2000, 182 (9): 2639-2642. 10.1128/JB.182.9.2639-2642.2000.
Oussenko IA, Sanchez R, Bechhofer DH: Bacillus subtilis YhaM, a member of a new family of 3'-to-5' exonucleases in gram-positive bacteria. J Bacteriol. 2002, 184 (22): 6250-6259. 10.1128/JB.184.22.6250-6259.2002.
Deutscher MP, Reuven NB: Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci USA. 1991, 88 (8): 3277-3280. 10.1073/pnas.88.8.3277.
Belasco JG, Higgins CF: Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988, 72 (1-2): 15-23. 10.1016/0378-1119(88)90123-0.
Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM: Copurification of E. coli RNase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994, 76 (5): 889-900. 10.1016/0092-8674(94)90363-8.
Miczak A, Kaberdin VR, Wei CL, Lin-Chao S: Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci USA. 1996, 93 (9): 3865-3869. 10.1073/pnas.93.9.3865.
Py B, Higgins CF, Krisch HM, Carpousis AJ: A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996, 381 (6578): 169-172. 10.1038/381169a0.
Lin PH, Lin-Chao S: RhlB helicase rather than enolase is the beta-subunit of the Escherichia coli polynucleotide phosphorylase (PNPase)-exoribonucleolytic complex. Proc Natl Acad Sci USA. 2005, 102 (46): 16590-16595. 10.1073/pnas.0500994102.
Lin-Chao S, Chiou NT, Schuster G: The PNPase, exosome and RNA helicases as the building components of evolutionarily-conserved RNA degradation machines. J Biomed Sci. 2007, 14 (4): 523-532. 10.1007/s11373-007-9178-y.
Kaberdin VR, Miczak A, Jakobsen JS, Lin-Chao S, McDowall KJ, von Gabain A: The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc Natl Acad Sci USA. 1998, 95 (20): 11637-11642. 10.1073/pnas.95.20.11637.
Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, Krisch HM, Carpousis AJ: Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998, 12 (17): 2770-2781. 10.1101/gad.12.17.2770.
Bernstein JA, Lin PH, Cohen SN, Lin-Chao S: Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc Natl Acad Sci USA. 2004, 101 (9): 2758-2763. 10.1073/pnas.0308747101.
Khemici V, Carpousis AJ: The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol Microbiol. 2004, 51 (3): 777-790. 10.1046/j.1365-2958.2003.03862.x.
Liou GG, Jane WN, Cohen SN, Lin NS, Lin-Chao S: RNA degradosomes exist in vivo in Escherichia coli as multicomponent complexes associated with the cytoplasmic membrane via the N-terminal region of ribonuclease E. Proc Natl Acad Sci USA. 2001, 98 (1): 63-68. 10.1073/pnas.011535498.
Leroy A, Vanzo NF, Sousa S, Dreyfus M, Carpousis AJ: Function in Escherichia coli of the non-catalytic part of RNase E: role in the degradation of ribosome-free mRNA. Mol Microbiol. 2002, 45 (5): 1231-1243. 10.1046/j.1365-2958.2002.03104.x.
Lopez PJ, Marchand I, Joyce SA, Dreyfus M: The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol Microbiol. 1999, 33 (1): 188-199. 10.1046/j.1365-2958.1999.01465.x.
Xu F, Cohen SN: RNA degradation in Escherichia coli regulated by 3' adenylation and 5' phosphorylation. Nature. 1995, 374 (6518): 180-183. 10.1038/374180a0.
Coburn GA, Mackie GA: Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J Mol Biol. 1998, 279 (5): 1061-1074. 10.1006/jmbi.1998.1842.
Carpousis AJ: The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007, 61: 71-87. 10.1146/annurev.micro.61.080706.093440.
Erce MA, Low JK, March PE, Wilkins MR, Takayama KM: Identification and functional analysis of RNase E of Vibrio angustum S14 and two-hybrid analysis of its interaction partners. Biochim Biophys Acta. 2009, 1794 (8): 1107-1114.
Chandran V, Luisi BF: Recognition of enolase in the Escherichia coli RNA degradosome. J Mol Biol. 2006, 358 (1): 8-15. 10.1016/j.jmb.2006.02.012.
Nurmohamed S, McKay AR, Robinson CV, Luisi BF: Molecular recognition between Escherichia coli enolase and ribonuclease E. Acta Crystallogr D Biol Crystallogr. 2010, 66 (Pt 9): 1036-1040. 10.1107/S0907444910030015.
Ait-Bara S, Carpousis AJ: Characterization of the RNA degradosome of Pseudoalteromonas haloplanktis: conservation of the RNase E-RhlB interaction in the {gamma}-Proteobacteria. J Bacteriol. 2010, 192 (20): 5413-5423. 10.1128/JB.00592-10.
Purusharth RI, Klein F, Sulthana S, Jager S, Jagannadham MV, Evguenieva-Hackenberg E, Ray MK, Klug G: Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J Biol Chem. 2005, 280 (15): 14572-14578. 10.1074/jbc.M413507200.
Cheng ZF, Deutscher MP: An important role for RNase R in mRNA decay. Mol Cell. 2005, 17 (2): 313-318. 10.1016/j.molcel.2004.11.048.
Hardwick SW, Chan VS, Broadhurst RW, Luisi BF: An RNA degradosome assembly in Caulobacter crescentus. Nucleic Acids Res. 2010
Banerjee S, Nandyala AK, Raviprasad P, Ahmed N, Hasnain SE: Iron-dependent RNA-binding activity of Mycobacterium tuberculosis aconitase. J Bacteriol. 2007, 189 (11): 4046-4052. 10.1128/JB.00026-07.
Jager S, Fuhrmann O, Heck C, Hebermehl M, Schiltz E, Rauhut R, Klug G: An mRNA degrading complex in Rhodobacter capsulatus. Nucl Acids Res. 2001, 29 (22): 4581-4588. 10.1093/nar/29.22.4581.
Lee K, Cohen SN: A Streptomyces coelicolor functional orthologue of Escherichia coli RNase E shows shuffling of catalytic and PNPase-binding domains. Mol Microbiol. 2003, 48 (2): 349-360. 10.1046/j.1365-2958.2003.03435.x.
Kovacs L, Csanadi A, Megyeri K, Kaberdin VR, Miczak A: Mycobacterial RNase E-associated proteins. Microbiol Immunol. 2005, 49 (11): 1003-1007.
McDowall KJ, Cohen SN: The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA-binding site. J Mol Biol. 1996, 255 (3): 349-355. 10.1006/jmbi.1996.0027.
Condon C: What is the role of RNase J in mRNA turnover?. RNA Biol. 2010, 7 (3): 10.4161/rna.7.3.11913.
Kime L, Jourdan SS, Stead JA, Hidalgo-Sastre A, McDowall KJ: Rapid cleavage of RNA by RNase E in the absence of 5' monophosphate stimulation. Mol Microbiol. 2010, 76 (3): 590-604. 10.1111/j.1365-2958.2009.06935.x.
Andrade JM, Hajnsdorf E, Regnier P, Arraiano CM: The poly(A)-dependent degradation pathway of rpsO mRNA is primarily mediated by RNase R. RNA. 2009, 15 (2): 316-326. 10.1261/rna.1197309.
Ghosh S, Deutscher MP: Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci USA. 1999, 96 (8): 4372-4377. 10.1073/pnas.96.8.4372.
Cohen SN: Surprises at the 3' end of prokaryotic RNA. Cell. 1995, 80 (6): 829-832. 10.1016/0092-8674(95)90284-8.
Dreyfus M, Regnier P: The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002, 111 (5): 611-613. 10.1016/S0092-8674(02)01137-6.
Xu F, Lin-Chao S, Cohen SN: The Escherichia coli pcnB gene promotes adenylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc Natl Acad Sci USA. 1993, 90: 6756-6760. 10.1073/pnas.90.14.6756.
Coburn GA, Mackie GA: Differential sensitivities of portions of the mRNA for ribosomal protein S20 to 3'-exonucleases dependent on oligoadenylation and RNA secondary structure. J Biol Chem. 1996, 271 (26): 15776-15781. 10.1074/jbc.271.26.15776.
Coburn GA, Mackie GA: Overexpression, purification, and properties of Escherichia coli ribonuclease II. J Biol Chem. 1996, 271 (2): 1048-1053. 10.1074/jbc.271.2.1048.
Hajnsdorf E, Regnier P: E. coli rpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J Mol Biol. 1999, 286 (4): 1033-1043. 10.1006/jmbi.1999.2547.
Haugel-Nielsen J, Hajnsdorf E, Regnier P: The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleolytic processing and exonucleolytic degradation. EMBO J. 1996, 15 (12): 3144-3152.
Kaberdin VR, Chao YH, Lin-Chao S: RNase E cleaves at multiple sites in bubble regions of RNA I stem loops yielding products that dissociate differentially from the enzyme. J Biol Chem. 1996, 271 (22): 13103-13109. 10.1074/jbc.271.22.13103.
Regnier P, Hajnsdorf E: Poly(A)-assisted RNA decay and modulators of RNA stability. Prog Mol Biol Transl Sci. 2009, 85: 137-185. full_text.
Lalonde MS, Zuo Y, Zhang J, Gong X, Wu S, Malhotra A, Li Z: Exoribonuclease R in Mycoplasma genitalium can carry out both RNA processing and degradative functions and is sensitive to RNA ribose methylation. RNA. 2007, 13 (11): 1957-1968. 10.1261/rna.706207.
Cao GJ, Sarkar N: Identification of the gene for an Escherichia coli poly(A) polymerase. Proc Natl Acad Sci USA. 1992, 89 (21): 10380-10384. 10.1073/pnas.89.21.10380.
Portnoy V, Schuster G: Mycoplasma gallisepticum as the first analyzed bacterium in which RNA is not polyadenylated. FEMS Microbiol Lett. 2008, 283 (1): 97-103. 10.1111/j.1574-6968.2008.01157.x.
Mechold U, Fang G, Ngo S, Ogryzko V, Danchin A: YtqI from Bacillus subtilis has both oligoribonuclease and pAp-phosphatase activity. Nucleic Acids Res. 2007, 35 (13): 4552-4561. 10.1093/nar/gkm462.
Fang M, Zeisberg WM, Condon C, Ogryzko V, Danchin A, Mechold U: Degradation of nanoRNA is performed by multiple redundant RNases in Bacillus subtilis. Nucleic Acids Res. 2009, 37 (15): 5114-5125. 10.1093/nar/gkp527.
Celesnik H, Deana A, Belasco JG: Initiation of RNA decay in Escherichia coli by 5' pyrophosphate removal. Mol Cell. 2007, 27 (1): 79-90. 10.1016/j.molcel.2007.05.038.
Acknowledgements
We thank Dr H Kuhn for editing of the manuscript. VRK was supported by IKERBASQUE (Basque Foundation for Science) and the Thematic Research Program of Academia Sinica (AS 97-23-22). DS was supported by Academia Sinica, Distinguished Postdoctoral Fellowship program. This work was also supported by grants from the National Science Council, Taiwan (NSC 98-2321-B-001-009; NSC 99-2321-B-001-004) and by an intramural fund from Academia Sinica to S L-C. We apologize to those authors whose work could not be cited due to space constraints.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
The manuscript was prepared by VRK, DS and SL-C. All authors read and approved the final manuscript.
Vladimir R Kaberdin, Dharam Singh contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kaberdin, V.R., Singh, D. & Lin-Chao, S. Composition and conservation of the mRNA-degrading machinery in bacteria. J Biomed Sci 18, 23 (2011). https://doi.org/10.1186/1423-0127-18-23
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1423-0127-18-23