Abstract
Background
Leg weakness issues are a great concern for the pig breeding industry, especially with regard to animal welfare. Traits associated with leg weakness are partly influenced by the genetic background of the animals but the genetic basis of these traits is not yet fully understood. The aim of this study was to identify quantitative trait loci (QTL) affecting leg weakness in pigs.
Methods
Three hundred and ten F2 pigs from a Duroc × Pietrain resource population were genotyped using 82 genetic markers. Front and rear legs and feet scores were based on the standard scoring system. Osteochondrosis lesions were examined histologically at the head and the condylus medialis of the left femur and humerus. Bone mineral density, bone mineral content and bone mineral area were measured in the whole ulna and radius bones using dual energy X-ray absorptiometry. A line-cross model was applied to determine QTL regions associated with leg weakness using the QTL Express software.
Results
Eleven QTL affecting leg weakness were identified on eight autosomes. All QTL reached the 5% chromosome-wide significance level. Three QTL were associated with osteochondrosis on the humerus end, two with the fore feet score and two with the rear leg score. QTL on SSC2 and SSC3 influencing bone mineral content and bone mineral density, respectively, reached the 5% genome-wide significance level.
Conclusions
Our results confirm previous studies and provide information on new QTL associated with leg weakness in pigs. These results contribute towards a better understanding of the genetic background of leg weakness in pigs.
Similar content being viewed by others
Background
Leg weakness (LW) has a great impact on fitness and longevity of animals, which influences not only animal welfare but also production and reproduction performance. It has been shown that between 20 and 50% of boars completing performance tests are rejected as breeding animals because of LW problems [1]. Genetic correlations between LW-related traits and longevity in breeding sows have been reported and suggest that a better leg status would decrease involuntary culling [2, 3]. Heritability estimates have been reported for LW in Duroc, Landrace, and Yorkshire sires i.e. 0.23, 0.30 and 0.39, respectively [4], and for an overall leg score in Landrace and Large White sows, i.e. 0.27 and 0.38, respectively [2]. In addition, osteochondrosis (OC) is regarded as the main cause of LW in pig [5, 6]. OC is a skeletal disease characterized by disturbed bone formation, cartilage retention, or necrosis of the cartilage canal in articular cartilage [7, 8] and results in economic losses mainly due to the culling of pigs in the breeding industry [9]. The disease occurs at high frequencies in growing pigs in all commercial breeds [10]. The estimated heritability of OC ranges from 0.06 to 0.5 [2, 5, 11, 12] in different pig breeds. Moreover, OC is reported to have negative effects on important performance traits such as sow longevity, growth and feed conversion rate [12, 13].
In addition to OC, bone mineral density (BMD) is generally regarded as an important parameter to assess bone growth and is associated with bone fracture risk and structural soundness in pigs. Studies in humans have shown that variation in BMD can be explained by genetic factors [14, 15]. Taken together, all the data reported so far imply that LW-related traits have a low to moderate heritability. Nevertheless, genetic studies of LW-related traits in growing and finishing pigs are limited. A significant number of QTL for performance traits has been reported in pigs [16] but few studies have been devoted to LW-related traits [17–21]. Therefore, the aim of this study was to investigate QTL for LW-related traits, including leg and feet scores, OC and bone mineral traits in a Duroc × Pietrain resource population.
Methods
Experimental animal population
In this study, we used 310 F2 pigs from a Duroc × Pietrain resource population comprising three generations, Parent (P), F1, and F2 pigs, and which had been previously analysed to detect QTL for growth, carcass and meat quality traits [22]. The F2 pigs were generated by mating six F1 males with 25 F1 females. All animals were maintained at the Frankenforst experimental research farm at the University of Bonn. Piglets were weaned at 28 days of age, males were castrated prior to weaning and placed in pens in the post-weaning unit until 10 weeks of age. The F2 pigs were given an ad libitum diet during the whole test period and were slaughtered at approximately 105 kg live weight at around 25-26 weeks of age in the slaughterhouse of the research farm Schwarzenau in Bavaria, Germany. Tissue samples from the tail were collected within the first week after birth for DNA isolation.
Phenotyping
Before slaughter, legs and feet were scored by the same person, using the criteria listed in additional file 1, Table S1 as guidelines to make assessments. The traits were recorded according to the rules of German performance stations [23]. Each 'leg score' is an assessment of the strength and straightness of the legs and of the stability of the joints. Leg scores ranged from 1 to 5, the optimum level being 3. Data were then transformed into a desirability scale, by using the absolute value of the original scores after subtracting three scores (score 3 becomes 0 for optimum leg score, score 2 and 4 become 1 for moderate leg score, and score 1 and 5 become 2 for poor leg score). For feet, the angle and strength of feet/leg attachment, soundness of toes and weight distribution on toes were assessed and given a score value between 1 (poor) and 3 (good). Leg and feet scores were measured on pigs walking on a solid concrete floor. After slaughter, the left fore and rear legs were removed from the carcass to carry out histological examinations of OC lesions. As OC is a bilaterally symmetrical syndrome, it was decided to examine only the left legs. The recorded OC lesions were scored 1 to 4, 4 for normal and 3 to 1 for mildly to severely affected (Additional file 1, Figure 1). OC lesions were evaluated on the head of the humerus (HH), condylus medialis humeri (CMH), head of the femur (HF) and the condylus medialis femoris (CMF). The histological examination assessed cartilage thickness, cartilage degradation and the vessel structure of cartilage canals. The histological procedures that were used have been described by Laenoi et al. [24]. The number of animals with OC on different joints ranged between 274 and 279 (Table 1). A total of 1,108 samples (532 from castrated and 576 from female pigs) out of 1,240 samples were phenotyped. In addition, the whole ulna and radius bones from the left carcass were stripped of all surrounding tissues and the bone mineral-related traits (BMD, BMC and BMA) were examined using dual energy X-ray absorptiometry (DXA) [25]. In total 275 animals were phenotyped for the DXA traits (Table 2).
Genotyping
Markers used for genotyping were mainly selected from the USDA/MARC map (http://www.marc.usda.gov) and included 79 microsatellites and three biallelic markers. Marker order and genetic distances between markers are described in additional file 2, Table S2. Genotyping, electrophoresis, and allele determination were carried out with a LI-COR 4200 Automated Sequencer (DNA Analyzer, GENE reader 4200). Allele and genotyping errors were checked using Pedcheck software (v 1.1) [26]. In addition to the microsatellite markers, SNP in genes assumed to affect cartilage quality were included, i.e. SNP located in the COL10A1 and MMP3 genes. Sequences were obtained from GenBank (accession no AF222861 and FJ788664 for porcine COL10A1 and MMP3, respectively) and assays were designed to permit genotyping using a multiplex SNP genotyping platform (Beckman Coulter). The relative positions of the markers were assigned using the build, two-point and fixed options of CRIMAP software, version 2.4 [27]. Recombination units were converted into map distances using the Kosambi mapping function. Marker information content and segregation distortion were tested. A linkage map was constructed with a total length of 2588.7 cM and an average marker interval of 31.57 cM.
Statistical analysis
The data were analysed using the software package SAS® (v 9.2, SAS® Inc., CA, USA). Generalized linear models (PROC GLM) were used to identify the effects of sire, dam, age, sex, birth weight, daily weight gain, litter size, litter effect, parity, season, and of carcass weight and length at slaughter on the investigated traits (Additional file 3, Table S3).
F2 QTL interval mapping was performed using the web-based program QTL Express [28] available at http://qtl.cap.ed.ac.uk/. The basic QTL regression model used in the present study was:

where: yi = phenotype of the ith offspring; μ = overall mean; Fi = fixed effect of litter; β = regression coefficient on the covariate; covi = covariate of average daily gain for leg and feet score age for OC, and slaughter weight and carcass length for DXA; cai = additive coefficient of the ith individual at a putative QTL; cdi = dominance coefficient of the ith individual at a putative QTL; a = additive effect of the putative QTL; d = dominance effect of the putative QTL; and εi = residual error.
The regression model was fitted at 1-cM intervals along each chromosome and the F-value for the QTL effect was calculated at each point. Thresholds for chromosome-wide significance were determined by 1000 data permutations [29] for individual chromosomes. Significance at the 5% chromosome-wide (CW) level was considered suggestive, 1% CW was considered significant and significance at the 5% genome-wide (GW) level as highly significant. To derive GW significance levels from the chromosome-wide significance levels, the Bonferoni correction was applied [30]. Empirical 95% confidence intervals (CI) and flanking markers for estimated QTL positions were obtained by applying the bootstrap approach with 1000 re-samplings [27]. The percentage of phenotype variation explained by a QTL was calculated as:

where, MSR is the mean square of the reduced model without QTL effects and MSF is the mean square of the full model.
Results
Distributions and correlation of the traits
Descriptive statistics of LW-related traits are given in Tables 1 and 2. It is important to note that in this study the direction of a desirable score is the difference between leg and feet scores and OC scores. For leg score, a low value is desirable but for feet and OC scores a high value is desirable. A high percentage of animals showed moderate fore feet scores (FFS) (79.4%) and good rear feet scores (RFS) (54.5%). Only 9.0% and 1.3% of animals showed poor feet scores for fore and rear feet, respectively. For the fore leg score (FLS), 42.3% of animals had a score value of 2 and for the rear leg score (RLS), 54.8% of animals had a score value of 3. Few animals had very poor leg scores (4.8% for fore leg and 0.3% for rear leg). Phenotypic correlations among FLS, RLS, FFS and RFS were low to medium, ranging from 0.19 to 0.44 (Table 1). The percentage of severe OC lesions in the 1,108 cartilage samples was higher in the CMF of the knee joint compared to other joints. The CMH and HH of fore limbs had healthier scores than CMF and HF. Phenotypic correlations among OC scores were very low, ranging from -0.13 to 0.12 (Table 1). BMD and BMC were not significantly different between castrated male pigs and female pigs (Table 2). The phenotypic correlation between BMD and BMC was positive (r = 0.70, P < 0.01). Parity, carcass length, weight at slaughter, age and average daily gain had significant (P < 0.05) effects on the measured traits (Table S2). Parity, carcass length and average daily gain had significant (P < 0.05) effects on FLS but only average daily gain (ADG) had an effect on RLS. Parity showed effects on FFS, HH, CMH and HF. Age also had an effect on HF. Parity, carcass length and weight at slaughter affected all DXA traits. BMD and BMC were highly correlated (P < 0.01) with the animals' weight at slaughter (r = 0.54 and 0.71, respectively).
QTL for leg weakness-related traits
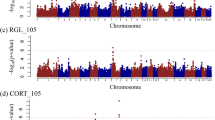
The results of the QTL analysis are given in Table 3. Eleven QTL were identified for LW-related traits on eight autosomes. Most QTL had highly significant dominance effects and three QTL were additive. Two chromosomal regions were identified for FFS (P ≤ 0.05, CW), at 166 cM on SSC1 and at 36 cM on SSC16. Two QTL, at 87 cM (P ≤ 0.05, CW) on SSC6 and at 26 cM on SSC18, were identified for RLS. No QTL was found for rear feet score and fore leg score. QTL associated with OC were located on SSC2, 3, 6, 10 and 14. The OC score of HH was influenced by three QTL regions, on SSC2, 3, and 6 at 14, 13 and 61 cM, respectively. A QTL for CMH was identified at 0 cM on SSC14. One QTL affecting OC score of CMF was identified on SSC10 at 70 cM. However, no suggestive QTL was found for OC score of HF. Two QTL were identified for bone mineral-related traits, one for BMD and one for BMC. A QTL for BMD was found on SSC3 at 71 cM. Only one QTL was detected for BMC, at 0 cM on SSC2. Both QTL for BMD and BMC reached a 5% GW significance.
In this study, most of the detected QTL appeared to have effects on only one trait, showing no effects on other traits. However, some chromosomal regions influenced more than one trait, notably on SSC2, 3 and 6.
Discussion
In this study, we evaluated conformation traits describing leg and feet condition, osteochondrosis score and bone mineral density, which are important in selection to reduce the risk of leg weakness in pigs. However, the genetics of LW-related traits is complex [12, 31]. A number of factors are known to influence the development of LW, such as nutrition imbalance, high body weight, rapid growth rate, bone and joint diseases, bad body and leg structure, and mechanical stress [11, 13]. Moreover, it has been reported that the degree of LW and OC may be related to the breed and sex of animals [32]. However, in our study there was no effect of gender on LW-related traits, which implies that frequencies of LW and OC vary and depend on the genetic background of the animals [33]. It has been reported that the Duroc pure breed shows the highest incidence of OC compared to other European pig breeds (Pietrain, Landrace and Yorkshire) [32]. Our data suggest that the unfavourable QTL allele for OC originates from both Duroc and Pietrain breeds (i.e. two QTL originated from the Duroc and three from the Pietrain) (Table 3) and that in Duroc and Pietrain crossbred animals, the fore legs are less susceptible to OC than the rear legs. Andersson-Eklund et al. [17] have also reported lower OC incidences in the humerus than in the femur in a Wild boar × Large White population. In addition, our data show that the frequency of OC is high (31.05%) at CMF, which agrees with a previous report of 30.0% by Kadarmideen et al. [12].
QTL analyses for leg weakness and bone-related traits have been performed in different pig breeds, including Landrace purebred [34], White Duroc × Erhualian [19, 21], Large White × Meishan [20], Duroc × Landrace and Duroc × Large White crossbred [18], and Wild boar × Large White [17]. To the best of our knowledge, our study is the first to map QTL for LW-related traits in a Duroc and Pietrain intercross. We have identified 11 QTL some of which being novel and some confirming previous studies [17–21, 34], as described in the next section. However, large confidence regions were obtained in this experiment, which represents a common problem in QTL studies and hampers the comparison of QTL results and their interpretation in terms of causative genes, since large confidence intervals can contain many potential candidate genes [35].
In this study, a QTL for FFS was detected on SSC1 at 166 cM. QTL for the same trait have been reported at 89 cM in a Landrace purebred [34] and at 52 cM in a Large White × Meishan intercross [20] on the same chromosome. The dominant QTL for FFS found on SSC16 at 36 cM is close to a previously reported dominant QTL at 27 cM for rear leg score [19]. The QTL identified for RLS on SSC6 and SSC18 are new and do not overlap with any previous study. A QTL associated with rear leg score was observed on SSC6, close to marker SW193 (SSC6q2.1), where the gene for transforming growth factor-beta 1 (TGFβ1) is located [36]. This gene is an important candidate for LW-related traits since TGFβ1 is a potent regulator of cell proliferation and influences the size and shape of the limb [37]. We identified a QTL for the OC score at HH on SSC2 at 14 cM, while Christensen et al. [18] have reported QTL associated with cartilage thickening of the medial part of condylus humeri at 15 cM on the same chromosome. In addition, a QTL with dominance effect identified for the OC score at HH on SSC6 at 61 cM is located close to previously reported QTL for depression of the proximal edge of the radius at 51 cM [18] and for physis score at 75 cM [20]. QTL for HH on SSC3 at 13 cM and for CMH on SSC14 at 0 cM are new QTL (Figure 1). Interestingly, the QTL for CMF on SSC10 at 70 cM is close to a previously identified QTL regions at 75 cM for OC lesion in the subchondral bone of the medial part of condylus humeri and at 83 cM for fissure between cartilage and bone in pigs [18]. The QTL on SSC2 at 0 cM, close to marker SW2443 (SSC2p17), was the only QTL detected for BMC. One of the highest linkage associations, reaching a 5% GW significance, was found on SSC3 at 71 cM for BMD. A potential candidate gene in this chromosomal region is the follicle-stimulating hormone receptor (FSHR) gene, which directly regulates bone mass [15]. These QTL for BMC and BMD are novel and do not overlap with previously reported QTL.
Most of the identified QTL show large dominance effects rather than additive affects (Table 3). It is important to note that the transformation done on the leg score traits in this study did not change the identified QTL regions since the interval mapping results for these traits using the original score ranging from 1-5 or the scale 0-2 were the same. This implies that the transformation done on the leg score is not the reason for over-dominance in this experiment.
In another QTL study in the same population, 31 of 71 QTL for growth, fatness, leanness and meat quality traits have also shown high dominance effects [22], as well as QTL for immune traits [38]. Lee et al. [20] have also reported that most QTL for LW-related traits in a Large white × Meishan cross show dominance. In addition, using principal components analysis, Andersson-Eklund et al. [17] have identified a QTL for OC with a significant and large effect of over-dominance. Therefore, the results from this study and from previous studies reported in the literature [17–20, 34] suggest that dominance plays a role in the genetic control of LW-related traits.
Most of the traits analysed in this study are categorical rather than normally distributed. Previous studies have shown that the QTL analysis method [39] used is suitable for categorical traits, with little loss of power [19, 20]. The low heritability of these traits indicates that they may be complex traits and may be under a polygenic control primarily by non-additive gene action or affected by a major gene with Mendelian transmission [31]. In this study, most of the QTL were identified as single-trait regions. This could be explained by the low phenotypic correlations observed between the traits in the population.
Conclusions
This is the first study identifying QTL affecting leg weakness and its related traits in a fast growing cross bred pig population between the Duroc and Pietrain breeds. Multiple QTL were detected for leg and feet scores, implying that these traits are controlled by multiple genes and that information from more than one QTL must be incorporated in selection procedures. Our results reveal novel QTL regions on SSC2 for BMC, on SSC3 for HH, on SSC6 and SSC18 for RLS, and on SSC14 for CMH, and also support some previously reported QTL regions. Although confidence intervals are large, these results will help to fine-map and identify candidate genes in these QTL regions using additional markers or gene polymorphisms located in the identified regions for LW-related traits in pigs.
Abbreviations
- ADG:
-
average daily gain
- BMD:
-
bone mineral density
- BMC:
-
bone mineral content
- BMA:
-
bone mineral area
- QTL:
-
quantitative trait loci
- DXA:
-
dual energy X-ray absorptiometry
- LW:
-
leg weakness
- FLS:
-
fore leg score
- RLS:
-
rear leg score
- FFS:
-
fore feet score
- RFS:
-
rear feet score
- OC:
-
osteochondrosis
- HH:
-
head of the humerus
- CMH:
-
condylus medialis humeri
- HF:
-
head of the femur
- CMF:
-
condylus medialis femori
- DuPi:
-
Duroc × Pietrain resource population.
References
Webb AJ, Russell WS, Sales DI: Genetics of leg weakness in performance-tested boars. Anim Prod. 1983, 36: 117-130. 10.1017/S0003356100040010.
de Sevilla XF, Fabrega E, Tibau J, Casellas J: Genetic background and phenotypic characterization over two farrowings of leg conformation defects in Landrace and Large White sows. J Anim Sci. 2009, 87: 1606-1612. 10.2527/jas.2008-1200.
López-Serrano M, Reinsch N, Looft H, Kalm E: Genetic correlations of growth, backfat thickness and exterior with stayability in Large White and Landrace sows. Livest Prod Sci. 2000, 64: 121-131.
Huang S, Tsou H, Kan M, Lin W, Chi C: Genetic study on leg weakness and its relationship with economic traits in central tested boars in subtropical area. Livest Prod Sci. 1995, 44: 53-59. 10.1016/0301-6226(95)00063-Q.
Jorgensen B, Andersen S: Genetic parameters for osteochondrosis in Danish Landrace and Yorkshire boars and correlation with leg weakness and production traits. Anim Sci. 2000, 71: 427-434.
Lundeheim N: Genetic analysis of osteochondrosis and leg weakness in the Swedish pig progeny testing scheme. Acta Agric Scand. 1987, 37: 159-173. 10.1080/00015128709436552.
Ytrehus B, Carlson CS, Ekman S: Etiology and pathogenesis of osteochondrosis. Vet Pathol. 2007, 44: 429-448. 10.1354/vp.44-4-429.
Ytrehus B, Ekman S, Carlson CS, Teige J, Reinholt FP: Focal changes in blood supply during normal epiphyseal growth are central in the pathogenesis of osteochondrosis in pigs. Bone. 2004, 35: 1294-1306. 10.1016/j.bone.2004.08.016.
Hill M: Economic relevance, diagnosis, and countermeasures for degenerative joint disease (osteoarthrosis) and dyschondroplasia (osteochondrosis) in pigs. J Am Vet Med Assoc. 1990, 197: 254-259.
Uhlhorn H, Dalin G, Lundeheim N, Ekman S: Osteochondrosis in wild boar-Swedish Yorkshire crossbred pigs (F2 generation). Acta Vet Scand. 1995, 36: 41-53.
Jorgensen B, Nielsen B: Genetic parameters for osteochondrosis traits in elbow joints of crossbred pigs and relationships with production traits. Anim Sci. 2005, 81: 319-324. 10.1079/ASC41890319.
Kadarmideen HN, Schworer D, Ilahi H, Malek M, Hofer A: Genetics of osteochondral disease and its relationship with meat quality and quantity, growth, and feed conversion traits in pigs. J Anim Sci. 2004, 82: 3118-3127.
Stern S, Lundeheim N, Johansson K, Andersson K: Osteochondrosis and leg weakness in pigs selected for lean tissue growth rate. Livest Prod Sci. 1995, 44: 45-52. 10.1016/0301-6226(95)00056-Q.
Amin S, Riggs BL, Atkinson EJ, Oberg AL, Melton LJ, Khosla S: A potentially deleterious role of IGFBP-2 on bone density in aging men and women. J Bone Miner Res. 2004, 19: 1075-1083. 10.1359/JBMR.040301.
Xu ZR, Wang AH, Wu XP, Zhang H, Sheng ZF, Wu XY, Xie H, Luo XH, Liao EY: Relationship of age-related concentrations of serum FSH and LH with bone mineral density, prevalence of osteoporosis in native Chinese women. Clin Chim Acta. 2009, 400: 8-13. 10.1016/j.cca.2008.09.027.
Hu ZL, Reecy JM: Animal QTLdb: beyond a repository. A public platform for QTL comparisons and integration with diverse types of structural genomic information. Mamm Genome. 2007, 18: 1-4. 10.1007/s00335-006-0105-8.
Andersson-Eklund L, Uhlhorn H, Lundeheim N, Dalin G, Andersson L: Mapping quantitative trait loci for principal components of bone measurements and osteochondrosis scores in a wild boar × Large White intercross. Genet Res. 2000, 75: 223-230. 10.1017/S0016672399004371.
Christensen OF, Busch ME, Gregersen VR, Lund MS, Nielsen B, Vingborg RKK, Bendixen C: Quantitative trait loci analysis of osteochondrosis traits in the elbow joint of pigs. Animal. 2009, 4: 417-424. 10.1017/S1751731109991248.
Guo YM, Ai HS, Ren J, Wang GJ, Wen Y, Mao HR, Lan LT, Ma JW, Brenig B, Rothschild MF, Haley CS, Huang LS: A whole genome scan for quantitative trait loci for leg weakness and its related traits in a large F2 intercross population between White Duroc and Erhualian. J Anim Sci. 2009, 87: 1569-1575. 10.2527/jas.2008-1191.
Lee GJ, Archibald AL, Garth GB, Law AS, Nicholson D, Barr A, Haley CS: Detection of quantitative trait loci for locomotion and osteochondrosis-related traits in Large White × Meishan pigs. Anim Sci. 2003, 76: 155-156.
Mao H, Guo Y, Yang G, Yang B, Ren J, Liu S, Ai H, Ma J, Brenig B, Huang L: A genome-wide scan for quantitative trait loci affecting limb bone lengths and areal bone mineral density of the distal femur in a White Duroc × Erhualian F2 population. BMC Genet. 2008, 9: 63-10.1186/1471-2156-9-63.
Liu G, Jennen DG, Tholen E, Juengst H, Kleinwachter T, Holker M, Tesfaye D, Un G, Schreinemachers HJ, Murani E, Ponsuksili S, Kim JJ, Schellander K, Wimmer K: A genome scan reveals QTL for growth, fatness, leanness and meat quality in a Duroc-Pietrain resource population. Anim Genet. 2007, 38: 241-252. 10.1111/j.1365-2052.2007.01592.x.
Zentralverband der Deutschen Schweineproduktion (ZDS): Richtlinie für die Stationsprüfung auf Mastleistung, Schlachtkörperwert und Fleischbeschaffenheit beim Schwein, 10.12.2003. Book Richtlinie für die Stationsprüfung auf Mastleistung, Schlachtkörperwert und Fleischbeschaffenheit beim Schwein, 10.12.2003 Bonn.
Laenoi W, Uddin MJ, Cinar MU, Phatsara C, Tesfaye D, Scholz AM, Tholen E, Looft C, Mielenz M, Sauerwein H, Schellander K: Molecular characterization and methylation study of matrix gla protein in articular cartilage from pig with osteochondrosis. Gene. 2010, 459: 24-31. 10.1016/j.gene.2010.03.009.
Mitchell AD, Scholz AM, Pursel VG: Total body and regional measurements of bone mineral content and bone mineral density in pigs by dual energy X-ray absorptiometry. J Anim Sci. 2001, 79: 2594-2604.
O'Connell JR, Weeks DE: PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998, 63: 259-266.
Green P, Fall K, Crooks S: Documentation for CRIMAP, Version 2.4. Washington University School of Medicine, St Louis, MO. 1990
Seaton G, Haley CS, Knott SA, Kearsey M, Visscher PM: QTL Express: mapping quantitative trait loci in simple and complex pedigrees. Bioinformatics. 2002, 18: 339-340. 10.1093/bioinformatics/18.2.339.
Churchill GA, Doerge RW: Empirical threshold values for quantitative trait mapping. Genetics. 1994, 138: 963-971.
de Koning DJ, Rattink AP, Harlizius B, van Arendonk JA, Brascamp EW, Groenen MA: Genome-wide scan for body composition in pigs reveals important role of imprinting. Proc Natl Acad Sci USA. 2000, 97: 7947-7950. 10.1073/pnas.140216397.
Kadarmideen HN, Janss LL: Evidence of a major gene from Bayesian segregation analyses of liability to osteochondral diseases in pigs. Genetics. 2005, 171: 1195-1206. 10.1534/genetics.105.040956.
Van der Wal PG, Goedegebuure SA, Van der Valk PC, Engel B, Van Essen G: Leg weakness and osteochondrosis in pigs; differences between the sexes of four breeds. Livest Prod Sci. 1978, 16: 65-74. 10.1016/0301-6226(87)90027-3.
Rothschild MF, Christain LL: Genetic control of front-leg weakness in Duroc swine. I. Direct response to five generations of divergent selection. Livest Prod Sci. 1988, 19: 459-471. 10.1016/0301-6226(88)90012-7.
Uemoto Y, Sato S, Ohnishi C, Hirose K, Kameyama K, Fukawa K, Kudo O, Kobayashi E: Quantitative trait loci for leg weakness traits in a Landrace purebred population. Anim Sci J. 2010, 81: 28-33. 10.1111/j.1740-0929.2009.00713.x.
de Koning DJ, Carlborg O, Haley CS: The genetic dissection of immune response using gene-expression studies and genome mapping. Vet Immunol Immunopathol. 2005, 105: 343-352. 10.1016/j.vetimm.2005.02.007.
Yerle M, Archibald AL, Dalens M, Gellin J: Localization of the PGD and TGF beta-1 loci to pig chromosome 6q. Anim Genet. 1990, 21: 411-417. 10.1111/j.1365-2052.1990.tb01985.x.
Thorp BH, Ekman S, Jakowlew SB, Goddard C: Porcine osteochondrosis: Deficiencies in transforming growth factor-β and insulin-like growth factor-I. Calcif Tissue Int. 1995, 56: 376-381. 10.1007/BF00301606.
Uddin MJ, Cinar MU, Große-Brinkhaus C, Tesfaye D, Tholen E, Juengst H, Looft C, Wimmers K, Phatsara C, Schellander K: Mapping quantitative trait loci for innate immune response in the pig. Int J of Immunogenet. 2011, 38: 121-131. 10.1111/j.1744-313X.2010.00985.x.
Visscher PM, Haley CS, Knott SA: Mapping QTLs for binary traits in backcross and F2 populations. Genet Res. 1996, 68: 55-63. 10.1017/S0016672300033887.
Acknowledgements
This work was supported by the German Federal Ministry of Education and Research (BMBF), and was part of the cooperative project 'FUGATO-plus' (sub-project GENE-FL), grant nr. FK20315135C. We greatly appreciate the excellent sample supply from the experimental station 'Frankenforst'.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
WL performed OC phenotyping, analysed the phenotypes, prepared and drafted the manuscript. MU contributed to the data analyses, prepared and edited the manuscript. MC, CL and KW shared manuscript editing. CG calculated the genetic cards and helped with the statistical analysis. DT supervised the lab work. EJ and ET supervised the statistical analysis and edited the manuscript. AS analysed the DXA traits. HJ was responsible for animal breeding and for collecting leg and feet score phenotypes. HS and MM supervised the cartilage and bone collection and histological analyses of the OC trait. CP supervised the whole work and was included in project management and organisation of samples and work flow. KS supervised the study and edited the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12711_2011_2485_MOESM1_ESM.PDF
Additional file 1: Table S1 - Basis of scoring for legs, feet and osteochondrosis. criteria used in this study to determine leg, feet and osteochondrosis scores Figure S1 - Sample of histological templates for the evaluation of OC score OC lesions are classified into four score values: (1) massive alterations of the cartilage including necrotic or ossified areas, (2) severe changes in the surface and deeper area of the articular cartilage like surface erosion, fibrillations, hyperplasia and chondrocyte necrosis, (3) cartilage shows few changes in surface and fibrillation, (4) cartilage surface is smooth, the matrix and chondrocytes are well organized with only a marginally rough surface or a weakly eosinophilic matrix or fibrillation (PDF 1 MB)
12711_2011_2485_MOESM2_ESM.PDF
Additional file 2: Table S2 - Markers used in the QTL analysis and genetic map as established from the DuPi resource population. anumbers in brackets at the first and last marker are relative positions of those in the USDA-MARC v2 linkage map; bS0226 not covered by USDA-MARC v2, but SW14, which is closely linked to S0226 (PigMap v 1.5); cS0035 at 0 and S0003 at 144.5 cM in the International Workshop 1 SSC6 integrated map with a total length of 166.0 cM (PDF 17 KB)
12711_2011_2485_MOESM3_ESM.PDF
Additional file 3: Table S3 - Analysis of variance for different LW-related traits. 1FLS = fore leg score, RLS = rear leg score, FFS = fore feet score, RFS = rear feet score, OC = osteochondrosis, HH = head of the humerus, CMH = condylus medialis humeri, HF = head of the femur, CMF = condylus medialis femori, BMD = bone mineral density, BMC = bone mineral content, BMA = bone mineral area, ADG = average daily gain (PDF 15 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Laenoi, W., Uddin, M.J., Cinar, M.U. et al. Quantitative trait loci analysis for leg weakness-related traits in a Duroc × Pietrain crossbred population. Genet Sel Evol 43, 13 (2011). https://doi.org/10.1186/1297-9686-43-13
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1297-9686-43-13