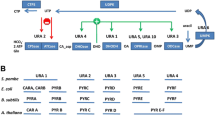
Abstract
Evolution of multigene families are considered in the review on the example of the PHO gene family encoding the structure of acid phosphatases in the yeast Saccharomyces cerevisiae. Analysis of the databases demonstrated that divergence due to a change in regulation of the structural genes and their inclusion in new regulatory networks is the main direction of evolution of multigene families encoding exoenzymes.
Similar content being viewed by others
References
Savinov, V.A., Sambuk, E.V., and Padkina, M.V., Natural and recombinant phytases of microorganisms, Vestnik SPbU, vol. 3, no. 2, pp. 66–75.
Abdulrehman, D., Monteiro, P.T., Teixeira, M.C., et al., YEASTRACT: providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface, Nucl. Acids Res., 2011, vol. 39, pp. D136–D140.
Albertin, W. and Marullo, P., Polyploidy in fungi: evolution after whole-genome duplication, Proc. Biol. Sci., 2012, vol. 279, pp. 497–509.
Almer, A. and Horz, W., Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast, EMBO J., 1986, vol. 5, pp. 2681–2687.
Bergman, L.W., Stranathan, M.C., and Preis, L.H., Structure of the transcriptionally repressed phosphaterepressible acid phosphatase gene (PHO5) of Saccharomyces cerevisiae, Mol. Cell. Biol., 1986, vol. 6, pp. 38–46.
Carlson, M., Celenza, J.L., and Eng, F.J., Evolution of the dispersed SUC gene family of Saccharomyces by rearrangements of chromosome telomeres, Mol. Cell. Biol., 1985, vol. 5, pp. 2894–2902.
Chatr-Aryamontri, A., Breitkreutz, B.-J., Heinicke, S., et al., The BioGR ID interaction database: 2013 update, Nucleic Acids Res., 2013, vol. 41 (Database issue), pp. D816–D823. doi: 10.1093/nar/gks1158
Christiaens, J.F., Van Mulders, S.E., Duitama, J., et al., Functional divergence of gene duplicates through ectopic recombination, EMBO Rep., 2012, vol. 13, pp. 1145–1151.
Cliften, P.F., Fulton, R.S., Wilson, R.K., and Johnston, M., After duplication: gene loss and adaptation in Saccharomyces genome, Genetics, 2006, vol. 172, pp. 863–872.
De Steensma, H.Y., de Jonge, P., Kaptein, A., and Kaback, D.B., Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: localization of a repeated sequence containing an acid phosphatase gene near a telomere of chromosome I and chromosome VIII, Curr. Genet., 1989, vol. 16, pp. 131–137.
Dong, D., Yuan, Z., and Zhang, Z., Evidences for increased expression variation of duplicate genes in budding yeast: from cisto trans-regulation effects, Nucleic Acids Res., 2011, vol. 39, pp. 837–847.
Fares, M.A., Keane, O.M., Toft, C., et al., The roles of whole-genome and small-scale duplications in the functional specialization of Saccharomyces cerevisiae genes, PLoS Genet., 2013, vol. 9, p. e1003176. doi: 10.1371/journal.pgen.1003176
Franceschini, A., Szklarczyk, D., Frankild, S., et al., STR ING v9.1: protein-protein interaction networks, with increased coverage and integration, Nucleic Acids Res., 2013, vol. 1 (Database issue), pp. D808–D815.
Force, A., Lynch, M., Pickett, F.B., et al., Preservation of duplicate genes by complementary, degenerative mutations, Genetics, 1999, vol. 151, pp. 1531–1545.
Gregory, P.D., Schmid, A., Zavari, M., Lui, L., Berger, S.L., and Horz, W., Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast, Mol. cell, 1998, Vol. 1, pp. 495–505.
Harbison, C.T., Gordon, D.B., Lee, T.I., et al., Transcriptional regulatory code of an eukaryotic genome, Nature, 2004, vol. 431, pp. 99–104.
Hu, Z., Killion, P.J., and Iyer, V.R., Genetic reconstruction of a functional transcriptional regulatory network, Nat. Genet., 2007, vol. 39, pp. 683–687.
Hurles, M., Gene duplication: the genomic trade in spare parts, PLoS Biol., 2004, vol. 2, p. E206.
Katju, V., Farslow, J.C., and Bergthorsson, U., Variation in gene duplicates with low synonymous divergence in Saccharomyces cerevisiae relative to Caenorhabditis elegans, Genome Biol., 2009, vol. 10, p. R75. doi: 10.1186/gb-2009-10-7-r75
Kowalska, E. and Kozik, A., The genes and enzymes involved in the biosynthesis of thiamin and thiamin diphosphate in yeasts, Cell. Mol. Biol. Lett., 2008, vol. 13, pp. 271–282.
Kroll, K., Pahtz, V., and Kniemeyer, O., Elucidating the fungal stress response by proteomics. J. Proteomics, 2013, vol. 10.
Lau, W., Schneider, K.R., and O’Shea, E.K., A genetic study of signaling processes for repression of PHO5 transcription in Saccharomyces cerevisiae, Genetics, 1998, vol. 150, pp. 1349–1359.
Lau, W.-T., Howson, R.W., Malkus, P., et al., Pho86p, an endoplasmic reticulum (ER) resident protein in Saccharomyces cerevisiae is required for ER exit of the high-affinity phosphate transporter Pho84p, Proc. Natl. Acad. Sci. USA, 2000, vol. 97, pp. 1107–1112
Levasseur, A. and Pontarotti, P., The role of duplications in the evolution of genomes highlights the need for evolutionary-based approaches in comparative genomics, Biology Direct, 2011, vol. 6, pp. 11–23.
Li, H. and Johnson, A.D., Evolution of transcription networks-lessons from yeasts, Curr. Biol., 2010, vol. 20, pp. 746–753.
Lynch, M. and Conery, J.S., The evolutionary fate and consequences of duplicate genes, Science, 2000, vol. 290, pp. 1151–1155.
Mao, C., Brown, C.R., Griesenbeck, J., and Boeger, H., Occlusion of regulatory sequences by promoter nucleosomes in vivo, PLoS One, 2011, vol. 6, p. e17521. doi:10.1371/journal.pone.0017521
Meyhack, B., Baiwa, W., Rudolph, H., and Hinnen, A., Two yeast acid phosphatase structural genes are the result of a tandem duplication and show different degrees of homology in their promoter and coding sequences, EMBO J., 1982, vol. 1, pp. 675–680.
Mizunaga, T., Izawa, M., Ikeda, K., et al., Secretion of an active nonglycosylated form of the repressible acid phosphatase of Saccharomyces cerevisiae in the presence of tunicamycin at low temperatures, J. Biochem. (Tokyo), 1988, vol. 103, pp. 321–326.
Nishimura, K., Yasumura, K., Igarashi, K., Harashima, S., and Kakinuma, Y., Transcription of some PHO genes in Saccharomyces cerevisiae is regulated by spt7p, Yeast, 1999, vol. 15, pp. 1711–1717.
Nosaka, K., Nishimura, H., and Iwashima, A., Identity of soluble thiamine-binding protein with thiamine repressible acid phosphatase in Saccharomyces cerevisiae, Yeast, 1989, vol. 5, pp. 447–451.
Ohno, S., Patterns in genome evolution, Curr. Opin. Genet. Dev., 1993, vol. 3, pp. 911–914.
Papp, B., Pal, C., and Hurst, L.D., Evolution of cis-regulatory elements in duplicated genes in yeast, Trends Genet., 2003, vol. 19, pp. 417–422.
Sambuk, E.V., Fizikova, A.Y., Savinov, V.A., and Padkina, M.V., Acid phosphatases of budding yeast as a model of choice for transcription regulation research, Enzyme Res., 2011, vol. 2011, p. 356093. doi: 10.4061/2011/356093
Shnyreva, M.G., Petrova, E.V., Egorov, S.N., and Hinnen, A., Biochemical properties and excretion behavior of repressible acid phosphatases with altered subunit composition, Microbiol. Res., 1996, vol. 151, pp. 291–300.
Singleton, C.K., Identification and characterization of the thiamine transporter gene of Saccharomyces cerevisiae, Gene, 1997, vol. 15, pp. 111–121.
Takashita, H., Kajiwara, Y., Shimoda, M., et al., Genetic instability of constitutive acid phosphatase in shochu and sake yeast, J. Biosci. Bioeng., 2013, vol. 116, pp. 71–78.
Thill, G.P., Kramer, R.A., Turner, K.J., and Bostian, K.A., Comparative analysis of the 5’-end regions of two repressible acid phosphatase genes in Saccharomyces cerevisiae, Mol. Cell. Biol., 1983, vol. 3, pp. 570–579.
Kakimoto, S., Genes coding for the structure of the acid phosphatases in Saccharomyces cerevisiae, Mol. Gen. Genet., 1975, vol. 143, pp. 65–70.
Tsai, Z.T., Tsai, H.K., Cheng, J.H., et al., Evolution of cisregulatory elements in yeast de novo and duplicated new genes, BMC Genomics, 2012, vol. 13, pp. 717–729.
Van Hoek, M.J. and Hogeweg, P., Metabolic adaptation after whole genome duplication, Mol. Biol. Evol., 2009, vol. 26, pp. 2441–2453.
Venter, U. and Hörz, W., The acid phosphatase genes PHO10 and PHO11 in S. cerevisiae are located at the telomeres of chromosomes VIII and I, Nucleic Acids Res., 1989, vol. 17, pp. 1353–1369
Yona, A.H., Manor, Y.S., Herbst, R.H., et al., Chromosomal duplication is a transient evolutionary solution to stress, Proc. Natl. Acad. Sci. USA, 2012, vol. 109, pp. 21010–21015.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © E.V. Sambuk, M.V. Padkina, 2013, published in Ekologicheskaya Genetika, 2013, Vol. 11, No. 4, pp. 34–44.
Rights and permissions
About this article
Cite this article
Sambuk, E.V., Padkina, M.V. Divergence of expression of PHO3, PHO5, PHO11, and PHO12 paralogs in the yeast Saccharomyces cerevisiae is a mechanism of evolution of multigene families. Russ J Genet Appl Res 5, 82–90 (2015). https://doi.org/10.1134/S2079059715020100
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S2079059715020100