Abstract
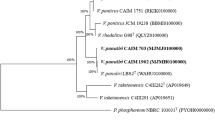
The genus Pseudomonas is one of the most diverse and ecologically important groups of bacteria. Numerous representatives of the genus are found in microbial communities of all natural environments, including those closely associated with plants and animals. This ubiquitous distribution determines a necessity of their physiological and genetic adaptations. Molecular methods revealed that bacteria of the genus Pseudomonas were predominant in ulcerative lesions on the skin of Baikal yellowfin Cottocomephorus grewingkii (Dybowski, 1874). According to ribosomal phylogeny, cultivated Pseudomonas spp. isolated from both ulcerative lesions and the water column of Lake Baikal were grouped into the intrageneric cluster IG P. fluorescens. The topology of the phylogenetic tree based on the gene for outer membrane porin OprF generally coincided with that based on the 16S rRNA genes at the intrageneric level; however, it reflected ecological features of the strains of the genus Pseudomonas at the subgroup level. Screening of pathogenicity determinants detected the oprL, ecfX, fliC, and algD genes in the genomes of Pseudomonas spp. isolated from the ulcerative lesions of fish, whereas oprL and gyrB genes were determined in the strains isolated from the water column.
Similar content being viewed by others
References
Peix, A., Berge, O., Rivas, R., et al., Pseudomonas argentinensis sp. nov., a novel yellow pigment-producing bacterial species, isolated from rhizospheric soil in Cordoba, Argentina, Int. J. Syst. Evol. Microbiol., 2005, vol. 55, no. 3, pp. 1107–1112. doi 10.1099/ijs.0.63445-0
Pavlova, O.N., Dryukker, V.V., Kostornova, T.Ya., and Nikulina, I.G., Pseudomonas bacteria distribution in Lake Baikal, Sib. Ekol. Zh., 2003, vol. 3, pp. 267–272.
Schwartz, T., Armant, O., Bretschneider, N., et al., Whole genome and transcriptome analyses of environmental antibiotic sensitive and multi-resistant Pseudomonas aeruginosa isolates exposed to waste water and tap water, Microbiol. Biotechnol., 2015, vol. 8, no. 1, pp. 116–130. doi 10.1111/1751-7915.12156
Mohanram, R., Jagtap, C., and Kumar, P., Isolation, screening, and characterization of surface-active agentproducing, oil-degrading marine bacteria of Mumbai Harbor, Mar. Pollut. Bull., 2016, vol. 105, pp. 131–138. doi 10.1016/j.marpolbul.2016.02.040
Bel’kova, N.L., Denikina, N.N., Sukhanova, E.V., et al., Heterogeneity of populations of organotrophic microorganisms on outer tegumental layers of sick fish, Voda: Khim. Ekol., 2016, no. 4 (94), pp. 32–39.
Nishimori, E., Kita-Tsukamoto, K., and Wakabayashi, H., Pseudomonas plecoglossicida sp. nov., the causative agent of bacterial haemorrhagic ascites of ayu, Plecoglossus altivelis, Int. J. Syst. Evol. Microbiol., 2000, vol. 50, no. 1, pp. 83–89. doi 10.1099/00207713-50-1-83
Saiman, L. and Siegel, J., Infection control in cystic fibrosis, Clin. Microbiol. Rev., 2004, vol. 17, pp. 57–71. doi 10.1128/CMR.17.1.57-71.2004
Reddy, G.S., Matsumoto, G.I., Schumann, P., et al., Psychrophilic pseudomonads from Antarctica: Pseudomonas antarctica sp. nov., Pseudomonas meridiana sp. nov. and Pseudomonas proteolytica sp. nov., Int. J. Syst. Evol. Microbiol., 2004, vol. 54, no. 3, pp. 713–719. doi 10.1099/ijs.0.02827-0
Yumoto, I., Yamazaki, K., Hishinuma, M., et al., Pseudomonas alcaliphila sp. nov., a novel facultatively psychrophilic alkaliphile isolated from seawater, Int. J. Syst. Evol. Microbiol., 2001, vol. 51, pp. 349–355. doi 10.1099/00207713-51-2-349
Zhong, Z.P., Liu, Y., Hou, T.T., et al., Pseudomonas salina sp. nov., isolated from a salt lake, Int. J. Syst. Evol. Microbiol., 2015, vol. 65, no. 9, pp. 2846–2851. doi 10.1099/ijs.0.000341
Moore, E.R.B., Mau, M., Arnscheidt, A., et al., The determination and comparison of the 16S rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of the natural intrageneric relationships, Syst. Appl. Microbiol., 1996, vol. 19, pp. 478–492. https://doi.org/10.1016/S0723-2020(96)80021-X.
Yamamoto, S., Kasai, H., Arnold, D.L., et al., Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes, Microbiology, 2000, vol. 146, pp. 2385–2394. doi 10.1099/00221287-146-10-2385
Mulet, M., Lalucat, J., and Garcia-Valdes, E., DNA sequence-based analysis of the Pseudomonas species, Environ. Microbiol., 2010, vol. 12, pp. 1513–1530. doi 10.1111/j.1462-2920.2010.02181.x
Bodilis, J., Nsigue Meilo, S., Cornelis, P., et al., A long-branch attraction artifact reveals an adaptive radiation in Pseudomonas, Mol. Biol. Evol., 2011, vol. 28, no. 10, pp. 2723–2726. doi 10.1093/molbev/msr099
Ullstrom, C.A., Siehnel, R., Woodruff, W., et al., Conservation of the gene for outer membrane protein OprF in the family Pseudomonadaceae: sequence of the Pseudomonas synringae oprF gene, J. Bacteriol., 1991, vol. 173, pp. 768–775. doi 10.1128/jb.173.2.768-775.1991
Bodilis, J. and Barray, S., Molecular evolution of the major outer membrane protein gene (oprF) of Pseudomonas, Microbiology, 2006, vol. 152, pp. 1075–1088. doi 10.1099/mic.0.28656-0
Bodilis, J., Hedde, M., Orange, N., and Barray, S., OprF polymorphism as a marker of ecological niche in Pseudomonas, Environ. Microbiol., 2006, vol. 8, no. 9, pp. 1544–1551. doi 10.1111/j.1462-2920.2006.01045.x
Bodilis, J., Calbrix, R., Guerillon, J., et al., Phylogenetic relationships between environmental and clinical isolates of Pseudomonas fluorescens and related species deduced from 16S rRNA gene and OprF protein sequences, Syst. Appl. Microbiol., 2004, vol. 27, pp. 93–108. doi 10.1078/0723-2020-00253
Lavenir, R., Jocktane, D., Laurent, F., et al., Improved reliability of Pseudomonas aeruginosa PCR detection by the use of the species-specific ecfX gene target, J. Microbiol. Methods, 2007, vol. 70, no. 1, pp. 20–29. doi 10.1016/j.mimet.2007.03.008
Matthijs, S., Coorevits, A., Gebrekidan, T.T., et al., Evaluation of oprI and oprL genes as molecular markers for the genus Pseudomonas and their use in studying the biodiversity of a small Belgian River, Res. Microbiol., 2013, vol. 164, no. 3, pp. 254–261. doi 10.1016/j.resmic. 2012.12.001
Kravtsova, L.S., Izhboldina, L.A., Khanaev, I.V., et al., Disturbances of the vertical zoning of green algae in the coastal part of the Listvennichnyi gulf of Lake Baikal, Dokl. Biol. Sci., 2012, vol. 447, nos. 1-6, pp. 350–352.
Denikina, N.N., Dzyuba, E.V., Bel’kova, N.L., et al., The first case of disease of the sponge Lubomirskia baicalensis: investigation of its microbiome, Biol. Bull. (Moscow), 2016, vol. 43, no. 3, pp. 263–270. https://doi.org/10.1134/S106235901603002X.
Khanaev, I.V., Dzyuba, E.V., Kravtsova, L.S., and Grachev, M.A., The effect of bloom of filamentous green algae on the reproduction of yellowfin sculpin Cottocomephorus grewingkii (Dybowski, 1874) (Cottoidae) during ecological crisis in Lake Baikal, Dokl. Biol. Sci., 2016, vol. 467, nos. 1-6, pp. 63–64.
Belykh, M.P., Sukhanova, E.V., and Bel’kova, N.L., Specific features of cultured heterotrophic microorganisms from the Lake Baikal littoral zone, Izv. Irkutsk. Gos. Univ.: Ser. Biol. Ekol., 2013, vol. 6, no. 3(1), pp. 20–26.
Bel’kova, N.L., Molecular and genetic methods for the analysis of microbial communities, in Raznoobrazie mikrobnykh soobshchestv vnutrennikh vodoemov Rossii: uchebno-metodicheskoe posobie (Diversity of Microbial Communities in Inland Water Bodies of Russia: Study Guide), 2009, pp. 53–63.
Sambrook, J., Fritsch, E.F., Maniatis, T., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor: Cold Spring Harbor Laboratory, 1989, vols. 1, 2, 3.
Brosius, J., Dull, T.J., Sleeter, D.D., and Noller, H.F., Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli, J. Mol. Biol., 1981, vol. 148, pp. 107–127. https://doi.org/10.1016/ 0022-2836(81)90508-8.
Hall, T.A., BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT, Nucleic Acids. Symp. Ser., 1999, vol. 41, pp. 95–98.
Garrity, G.M., Brenner, D.J., Krieg, N.R., et al., Bergey’s Manual of Systematic Bacteriology, vol. 2: The Proteobacteria, part B: The Gammaproteobacteria, New York: Springer-Verlag, 2005, 2nd ed. doi 10.1007/0-387-28022-7
Garrity, G.M., Brenner, D.J., Krieg, N.R., et al., Bergey’s Manual of Systematic Bacteriology, vol. 2: The Proteobacteria, part C: The Alpha-, Beta-, Delta-, and Epsilonproteobacteria, New York: Springer-Verlag, 2005. doi 10.1007/0-387-29298-5
López-Cortés, A., Schumann, P., Pukall, R., and Stackebrandt, E., Exiguobacterium mexicanum sp. nov. and Exiguobacterium artemiae sp. nov., isolated from the brine shrimp Artemia franciscana, Syst. Appl. Microbiol., 2006, vol. 29, no. 3, pp. 183–190. doi 10.1016/j.syapm.2005.09.007
Zhang, Y. and Qiu, S., Examining phylogenetic relationships of Erwinia and Pantoea species using whole genome sequence data, Antonie van Leeuwenhoek, 2015, vol. 108, no. 5, pp. 1037–1046. doi 10.1007/s10482-015-0556-6
Vos, P., Garrity, G., Jones, D., et al., Bergey’s Manual of Systematic Bacteriology, vol. 3: The Firmicutes, New York: Springer-Verlag, 2009. doi 10.1007/978-0-387-68489-5
Hilario, E., Buckley, T., and Young, J., Improved resolution on the phylogenetic relationships among Pseudomonas by the combined analysis of atpD, carA, recA and 16S rDNA, Antonie van Leeuwenhoek, 2004, vol. 86, pp. 51–64. doi 10.1023/B:ANTO.0000024910.57117.16
Khan, A.A. and Cerniglia, C.E., Detection of Pseudomonas aeruginosa from clinical and environmental samples by amplification of the exotoxin A gene using PCR, Appl. Environ. Microbiol., 1994, vol. 60, no. 10, pp. 3739–3745.
Watanabe, K., Nelson, J., Harayama, S., and Kasai, H., ICB database: the gyrB database for identification and classification of bacteria, Nucleic Acids Res., 2001, vol. 29, no. 1, pp. 344–345.
Lee, C.S. and Lee, J., Evaluation of new gyrB-based real-time PCR system for the detection of B. fragilis as an indicator of human-specific fecal contamination, J. Microbiol. Methods, 2010, vol. 82, pp. 311–318. doi 10.1016/j.mimet.2010.07.012
Qin, X., Emerson, J., Stapp, J., et al., Use of real time PCR with multiple targets to identify Pseudomonas aeruginosa and other nonfermenting gram-negative bacilli from patients with cystic fibrosis, J. Clin. Microbiol., 2003, vol. 41, no. 9, pp. 4312–4317. doi 10.1128/JCM.41.9.4312-4317.2003
Spangenberg, C., Heuer, T., Burger, C., and Tummler, B., Genetic diversity of flagellins of Pseudomonas aeruginosa, FEBS Lett., 1996, vol. 396, no. 4, pp. 213–217. doi 10.1016/0014-5793(96)01099-X
De Vos, D., Lim, A., Jr., Pirnay, J.P., et al., Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL, J. Clin. Microbiol., 1997, vol. 35, no. 6, pp. 1295–1299.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © N.L. Bel’kova, E.V. Dzyuba, E.S. Klimenko, I.V. Khanaev, N.N. Denikina, 2018, published in Genetika, 2018, Vol. 54, No. 5.
Rights and permissions
About this article
Cite this article
Bel’kova, N.L., Dzyuba, E.V., Klimenko, E.S. et al. Detection and Genetic Characterization of Bacteria of the Genus Pseudomonas from Microbial Communities of Lake Baikal. Russ J Genet 54, 514–524 (2018). https://doi.org/10.1134/S1022795418040038
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795418040038