Abstract
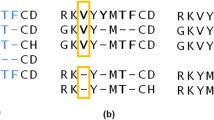
Verification of the PREFAB database containing golden standard protein alignments was performed. It has revealed a significant number of differences between the sequences from PREFAB and PDB databases. It was shown that, compared with the sequences given in the PDB, 575 alignments referred to a sequence with a gap; such alignments were excluded. Furthermore, compared with the PDB sequences, single substitutions or insertions were found for 440 amino acid sequences from PREFAB; these sequences were edited. SCOP domain analysis has shown that only 502 alignments in the resulting set contain sequences from the same family. Finally, eliminating duplicates, we have created a new golden standard alignment database PREFAB-P based on PREFAB; the PREFAB-P database contains 581 alignments.
Similar content being viewed by others
References
R. F. Smith and T. F. Smith, Protein Eng. 5, 35 (1992).
E. Deperieux, G. Baudoux, P. Briffeuil, et al., Comput. Appl. BioSci. 13, 249 (1997).
S. R. Eddy, ISMB 3, 114 (1995).
B. Morgenstern, A. Dress, and T. Werner, Proc. Natl. Acad. Sci. USA 93, 12098 (1996).
J. D. Thompson, T. J. Gibson, F. Plewniak, et al., Nucl. Acids Res. 24, 4876 (1997).
M. A. McClure, Vasi T. K., Fitch W. M., Mol. Biol. Evol. 11, 571 (1994).
M. R. Aniba, O. Poch, and J. D. Thompson, Nucl. Acids Res. 38, 7353 (2010).
R. C. Edgar, Nucl. Acids Res. 38, 2145 (2010).
R. C. Edgar, Nucl. Acids Res. 32, 1792 (2004).
A. G. Murzin, S. E. Brenner, T. Hubbard, and C. Chothia, J. Mol. Biol. 247, 536 (1995).
H. Hasegawa and L. Holm, Curr. Opin. Struct. Biol. 19, 341 (2009).
A. Godzik, Protein Sci. 5, 1325 (1996).
C. Etchebest, C. Benros, S. Hazout, and A. G. de Brevern, Proteins 59, 810 (2005).
C. Orengo, A. Michie, S. Jones, et al., Structure 5, 1093 (1997).
A. S. Siddiqui and G. J. Barton, Protein Sci. 42, 372 (1995).
M. B. Swindells, Protein Sci. 4, 103 (1995).
L. Holm and C. Sander, Proteins 19, 256 (1994).
A. Harrison, F. Pearl, R. Mott, et al., J. Mol. Biol. 5(323), 909 (2002).
F. M. Pearl, C. F. Bennett, J. E. Bray, et al., Nucl. Acids Res. 31, 452 (2003).
J. D. Thompson, F. Plewniak, and O. Poch, Bioinformatics 15, 87 (1999).
A. Bahr, J. D. Thompson, J. C. Thierry, and O. Poch, Nucl. Acids Res. 29, 323 (2001).
J. D. Thompson, P. Koehl, R. Ripp, and O. Poch, Proteins 61, 127 (2005).
E. Perrodou, C. Chica, O. Poch, et al., BMC Bioinformatics 9, 213 (2008).
K. Mizuguchi, C. M. Deane, T. L. Blundell, and J. P. Overington, Protein Sci. 7, 2469 (1998).
G. P. Raghava, S. M. Searle, P. C. Audley, et al., BMC Bioinformatics 4, 47 (2003).
I. Van Walle, I. Lasters, and L. Wyns, Bioinformatics 21, 1267 (2005).
H. M. Berman, K. Henrick, and H. Nakamura, Nat. Struct. Biol. 10, 980 (2003).
R. D. Finn, J. Mistry, J. Tate, et al., Nucl. Acids Res. 38, 211 (2010).
N. S. Boutonnet, M. J. Rooman, M. E. Ochagavia, et al., Protein Eng. 8, 647 (1995).
I. N. Shindyalov and P. E. Bourne, Protein Eng. 11, 739 (1998).
PREFAB v. 4.0: http://www.drive5.com/muscle/pre-fab.htm
R. Sadreyev and N. Grishin, J. Mol. Biol. 326, 317 (2003).
R. C. Edgar and K. A. Sjolander, Bioinformatics, DOI: 10.1093/bioinformatics/bth090 (2004).
R. C. Edgar and K. Sjolander, Bioinformatics, DOI: 10.1093/bioinformatics/bth091 (2004).
L. Holm and C. Sander, Nucl. Acids Res. 26, 316 (1998).
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © T.V. Astakhova, M.N. Lobanov, I.V. Poverennaya, M.A. Roytberg, V.V. Yacovlev, 2012, published in Biofizika, 2012, Vol. 57, No. 2, pp. 205–211.
Rights and permissions
About this article
Cite this article
Astakhova, T.V., Lobanov, M.N., Poverennaya, I.V. et al. Verification of the PREFAB alignment database. BIOPHYSICS 57, 133–137 (2012). https://doi.org/10.1134/S0006350912020030
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006350912020030