Abstract
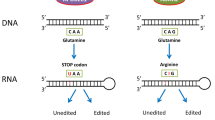
RNA editing by adenosine deaminases of the ADAR family attracts a growing interest of researchers, both zoologists studying ecological and evolutionary plasticity of invertebrates and medical biochemists focusing on the mechanisms of cancer and other human diseases. These enzymes deaminate adenosine residues in the double-stranded (ds) regions of RNA with the formation of inosine. As a result, some RNAs change their three-dimensional structure and functions. Adenosine-to-inosine editing in the mRNA coding sequences may cause amino acid substitutions in the encoded proteins. Here, we reviewed current concepts on the functions of two active ADAR isoforms identified in mammals (including humans). The ADAR1 protein, which acts non-specifically on extended dsRNA regions, is capable of immunosuppression via inactivation of the dsRNA interactions with specific sensors inducing the cell immunity. Expression of a specific ADAR1 splicing variant is regulated by the type I interferons by the negative feedback mechanism. It was shown that immunosuppressing effects of ADAR1 facilitate progression of some types of cancer. On the other hand, changes in the amino acid sequences resulting from the mRNA editing by the ADAR enzymes can result in the formation of neoantigens that can activate the antitumor immunity. The ADAR2 isoform acts on RNA more selectively; its function is associated with the editing of mRNA coding regions and can lead to the amino acid substitutions, in particular, those essential for the proper functioning of some neurotransmitter receptors in the central nervous system.
Similar content being viewed by others
Abbreviations
- ADAR:
-
adenosine deaminase, RNA-dependent
- dsRNA:
-
double-stranded RNA
- PKR:
-
protein kinase R
References
Lobas, A. A., Pyatnitskiy, M. A., Chernobrovkin, A. L., Ilina, I. Y., Karpov, D. S., Solovyeva, E. M., Kuznetsova, K. G., Ivanov, M. V., Lyssuk, E. Y., Kliuchnikova, A. A., Voronko, O. E., Larin, S. S., Zubarev, R. A., Gorshkov, M. V., and Moshkovskii, S. A. (2018) Proteogenomics of malignant melanoma cell lines: the effect of stringency of exome data filtering on variant peptide identification in shotgun proteomics, J. Proteome Res., 17, 1801–1811, doi: https://doi.org/10.1021/acs.jproteome.7b00841.
Kuznetsova, K. G., Kliuchnikova, A. A., Ilina, I. U., Chernobrovkin, A. L., Novikova, S. E., Farafonova, T. E., Karpov, D. S., Ivanov, M. V., Goncharov, A. O., Ilgisonis, E. V., Voronko, O. E., Nasaev, S. S., Zgoda, V. G., Zubarev, R. A., Gorshkov, M. V., and Moshkovskii, S. A. (2018) Proteogenomics of adenosine-to-inosine RNA editing in the fruit fly, J. Proteome Res., 17, 3889–3903, doi: https://doi.org/10.1021/acs.jproteome.8b00553.
Ishizuka, J. J., Manguso, R. T., Cheruiyot, C. K., Bi, K., Panda, A., Iracheta-Vellve, A., Miller, B. C., Du, P. P., Yates, K. B., Dubrot, J., Buchumenski, I., Comstock, D. E., Brown, F. D., Ayer, A., Kohnle, I. C., Pope, H. W., Zimmer, M. D., Sen, D. R., Lane-Reticker, S. K., Robitschek, E. J., Griffin, G. K., Collins, N. B., Long, A. H., Doench, J. G., Kozono, D., Levanon, E. Y., and Haining, W. N. (2019) Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade, Nature, 565, 43–48, doi: https://doi.org/10.1038/s41586-018-0768-9.
Orecchini, E., Doria, M., Antonioni, A., Galardi, S., Ciafre, S. A., Frassinelli, L., Mancone, C., Montaldo, C., Tripodi, M., and Michienzi, A. (2017) ADAR1 restricts LINE-1 retrotransposition, Nucleic Acids Res., 45, 155–168, doi: https://doi.org/10.1093/nar/gkw834.
Bass, B. L. (2002) RNA editing by adenosine deaminases that act on RNA, Annu. Rev. Biochem., 71, 817–846, doi: https://doi.org/10.1146/annurev.biochem.71.110601.135501.
Hough, R. F., and Bass, B. L. (1994) Purification of the Xenopus laevis double-stranded RNA adenosine deaminase, J. Biol. Chem., 269, 9933–9939.
Jin, Y., Zhang, W., and Li, Q. (2009) Origins and evolution of ADAR-mediated RNA editing, IUBMB Life, 61, 572–578, doi: https://doi.org/10.1002/iub.207.
Palladino, M. J., Keegan, L. P., O’Connell, M. A., and Reenan, R. A. (2000) dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing, RNA, 6, 1004–1018.
Melcher, T., Maas, S., Herb, A., Sprengel, R., Higuchi, M., and Seeburg, P. H. (1996) RED2, a brain-specific member of the RNA-specific adenosine deaminase family, J. Biol. Chem., 271, 31795–31798.
Oakes, E., Anderson, A., Cohen-Gadol, A., and Hundley, H. A. (2017) Adenosine deaminase that acts on RNA 3 (ADAR3) binding to glutamate receptor subunit B pre-mRNA inhibits RNA editing in glioblastoma, J. Biol. Chem., 292, 4326–4335, doi: https://doi.org/10.1074/jbc.M117.779868.
Schumacher, J. M., Lee, K., Edelhoff, S., and Braun, R. E. (1995) Distribution of Tenr, an RNA-binding protein, in a lattice-like network within the spermatid nucleus in the mouse, Biol. Reprod., 52, 1274–1283.
Saunders, L. R., and Barber, G. N. (2003) The dsRNA binding protein family: critical roles, diverse cellular functions, FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol., 17, 961–983, doi: https://doi.org/10.1096/fj.02-0958rev.
Herbert, A., Alfken, J., Kim, Y. G., Mian, I. S., Nishikura, K., and Rich, A. (1997 A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase, Proc. Natl. Acad. Sci. USA, 94, 8421–8426.
Nishikura, K., Yoo, C., Kim, U., Murray, J. M., Estes, P. A., Cash, F. E., and Liebhaber, S. A. (1991) Substrate specificity of the dsRNA unwinding/modifying activity, EMBO J., 10, 3523–3532.
Bass, B. L., and Weintraub, H. (1988) An unwinding activity that covalently modifies its double-stranded RNA substrate, Cell, 55, 1089–1098.
Licht, K., Janisiw, M. P., Jantsch, M. F., Anrather, D., Hartl, M., and Amman, F. (2018) Inosine induces context-dependent recoding and translational stalling, Nucleic Acids Res., 47, 3–14, doi: https://doi.org/10.1093/nar/gky1163.
Levanon, E. Y., Eisenberg, E., Yelin, R., Nemzer, S., Hallegger, M., Shemesh, R., Fligelman, Z. Y., Shoshan, A., Pollock, S. R., Sztybel, D., Olshansky, M., Rechavi, G., and Jantsch, M. F. (2004) Systematic identification of abundant A-to-I editing sites in the human transcriptome, Nat. Biotechnol., 22, 1001–1005, doi: https://doi.org/10.1038/nbt996.
Kim, D. D. Y., Kim, T. T. Y., Walsh, T., Kobayashi, Y., Matise, T. C., Buyske, S., and Gabriel, A. (2004) Widespread RNA editing of embedded alu elements in the human transcriptome, Genome Res., 14, 1719–1725, doi: https://doi.org/10.1101/gr.2855504.
Athanasiadis, A., Rich, A., and Maas, S. (2004) Wide-spread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome, PLoS Biol., 2, e391, doi: https://doi.org/10.1371/journal.pbio.0020391.
Blow, M., Futreal, P. A., Wooster, R., and Stratton, M. R. (2004) A survey of RNA editing in human brain, Genome Res., 14, 2379–2387, doi: https://doi.org/10.1101/gr.2951204.
Eisenberg, E., Nemzer, S., Kinar, Y., Sorek, R., Rechavi, G., and Levanon, E. Y. (2005) Is abundant A-to-I RNA editing primate-specific?, Trends Genet., 21, 77–81, doi: https://doi.org/10.1016/j.tig.2004.12.005..
Shen, M. R., Batzer, M. A., and Deininger, P. L. (1991) Evolution of the master Alu gene(s), J. Mol. Evol., 33, 311–320.
Garcia, M. A., Gil, J., Ventoso, I., Guerra, S., Domingo, E., Rivas, C., and Esteban, M. (2006) Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action, Microbiol. Mol. Biol. Rev., 70, 1032–1060, doi: https://doi.org/10.1128/MMBR.00027-06.
Yang, J.-H., Nie, Y., Zhao, Q., Su, Y., Pypaert, M., Su, H., and Rabinovici, R. (2003) Intracellular localization of differentially regulated RNA-specific adenosine deaminase isoforms in inflammation, J. Biol. Chem., 278, 45833–45842, doi: https://doi.org/10.1074/jbc.M308612200.
Clerzius, G., Gelinas, J.-F., Daher, A., Bonnet, M., Meurs, E. F., and Gatignol, A. (2009) ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication, J. Virol., 83, 10119–10128, doi: https://doi.org/10.1128/JVI.02457-08.
Porath, H. T., Knisbacher, B. A., Eisenberg, E., and Levanon, E. Y. (2017) Massive A-to-I RNA editing is common across the Metazoa and correlates with dsRNA abundance, Genome Biol., 18, 185, doi: https://doi.org/10.1186/s13059-017-1315-y.
Pestal, K., Funk, C. C., Snyder, J. M., Price, N. D., Treuting, P. M., and Stetson, D. B. (2015) Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development, Immunity, 43, 933–944, doi: https://doi.org/10.1016/j.immuni.2015.11.001.
Mannion, N. M., Greenwood, S. M., Young, R., Cox, S., Brindle, J., Read, D., Nellaker, C., Vesely, C., Ponting, C. P., McLaughlin, P. J., Jantsch, M. F., Dorin, J., Adams, I. R., Scadden, A. D. J., Ohman, M., Keegan, L. P., and O’Connell, M. A. (2014) The RNA-editing enzyme ADAR1 controls innate immune responses to RNA, Cell Rep., 9, 1482–1494, doi: https://doi.org/10.1016/j.celrep.2014.10.041.
Amberger, J. S., Bocchini, C. A., Schiettecatte, F., Scott, A. F., and Hamosh, A. (2015) OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders, Nucleic Acids Res., 43, D789–D798, doi: https://doi.org/10.1093/nar/gku1205.
Rice, G. I., Kasher, P. R., Forte, G. M. A., Mannion, N. M., Greenwood, S. M., Szynkiewicz, M., Dickerson, J. E., Bhaskar, S. S., Zampini, M., Briggs, T. A., Jenkinson, E. M., Bacino, C. A., Battini, R., Bertini, E., Brogan, P. A., Brueton, L. A., Carpanelli, M., De Laet, C., de Lonlay, P., del Toro, M., Desguerre, I., Fazzi, E., Garcia-Cazorla, A., Heiberg, A., Kawaguchi, M., Kumar, R., Lin, J. P., Lourenco, C. M., Male, A. M., Marques, W., Jr., Mignot, C., Olivieri, I., Orcesi, S., Prabhakar, P., Rasmussen, M., Robinson, R. A., Rozenberg, F., Schmidt, J. L., Steindl, K., Tan, T. Y., van der Merwe, W. G., Vanderver, A., Vassallo, G., Wakeling, E. L., Wassmer, E., Whittaker, E., Livingston, J. H., Lebon, P., Suzuki, T., McLaughlin, P. J., Keegan, L. P., O’Connell, M. A., Lovell, S. C., and Crow, Y. J. (2012) Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature, Nat. Genet., 44, 1243–1248, doi: https://doi.org/10.1038/ng.2414.
Miyamura, Y., Suzuki, T., Kono, M., Inagaki, K., Ito, S., Suzuki, N., and Tomita, Y. (2003) Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria, Am. J. Hum. Genet., 73, 693–699, doi: https://doi.org/10.1086/378209.
Samuel, C. E. (2011) Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral, Virology, 411, 180–193, doi: https://doi.org/10.1016/j.virol.2010.12.004.
Taylor, D. R., Puig, M., Darnell, M. E. R., Mihalik, K., and Feinstone, S. M. (2005) New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1, J. Virol., 79, 6291–6298, doi: https://doi.org/10.1128/JVI.79.10.6291-6298.2005.
Zahn, R. C., Schelp, I., Utermohlen, O., and von Laer, D. (2007) A-to-G hypermutation in the genome of lymphocytic choriomeningitis virus, J. Virol., 81, 457–464, doi: https://doi.org/10.1128/JVI.00067-06.
Suspene, R., Petit, V., Puyraimond-Zemmour, D., Aynaud, M.-M., Henry, M., Guetard, D., Rusniok, C., Wain-Hobson, S., and Vartanian, J.-P. (2011) Double-stranded RNA adenosine deaminase ADAR-1-induced hypermutated genomes among inactivated seasonal influenza and live attenuated measles virus vaccines, J. Virol., 85, 2458–2462, doi: https://doi.org/10.1128/JVI.02138-10.
Gandy, S. Z., Linnstaedt, S. D., Muralidhar, S., Cashman, K. A., Rosenthal, L. J., and Casey, J. L. (2007) RNA editing of the human herpesvirus 8 kaposin transcript eliminates its transforming activity and is induced during lytic replication, J. Virol., 81, 13544–13551, doi: https://doi.org/10.1128/JVI.01521-07.
Iizasa, H., Wulff, B.-E., Alla, N. R., Maragkakis, M., Megraw, M., Hatzigeorgiou, A., Iwakiri, D., Takada, K., Wiedmer, A., Showe, L., Lieberman, P., and Nishikura, K. (2010) Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency, J. Biol. Chem., 285, 33358–33370, doi: https://doi.org/10.1074/jbc.M110.138362.
Toth, A. M., Li, Z., Cattaneo, R., and Samuel, C. E. (2009) RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR, J. Biol. Chem., 284, 29350–29356, doi: https://doi.org/10.1074/jbc.M109.045146.
Gelinas, J.-F., Clerzius, G., Shaw, E., and Gatignol, A. (2011) Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA-activated protein kinase, J. Virol., 85, 8460–8466, doi: https://doi.org/10.1128/JVI.00240-11.
Casey, J. L. (2011) Control of ADAR1 editing of hepatitis delta virus RNAs, in Current Topics in Microbiology and Immunology, Vol. 353, pp. 123–143, doi: https://doi.org/10.1007/82_2011_146.
De Cecco, M., Ito, T., Petrashen, A. P., Elias, A. E., Skvir, N. J., Criscione, S. W., Caligiana, A., Brocculi, G., Adney, E. M., Boeke, J. D., Le, O., Beausejour, C., Ambati, J., Ambati, K., Simon, M., Seluanov, A., Gorbunova, V., Slagboom, P. E., Helfand, S. L., Neretti, N., and Sedivy, J. M. (2019) L1 drives IFN in senescent cells and promotes age-associated inflammation, Nature, 566, 73–78, doi: https://doi.org/10.1038/s41586-018-0784-9.
Gannon, H. S., Zou, T., Kiessling, M. K., Gao, G. F., Cai, D., Choi, P. S., Ivan, A. P., Buchumenski, I., Berger, A. C., Goldstein, J. T., Cherniack, A. D., Vazquez, F., Tsherniak, A., Levanon, E. Y., Hahn, W. C., and Meyerson, M. (2018) Identification of ADAR1 adenosine deaminase dependency in a subset of cancer cells, Nat. Commun., 9, 5450, doi: https://doi.org/10.1038/s41467-018-07824-4.
Xu, L.-D., and Ohman, M. (2018) ADAR1 editing and its role in cancer, Genes (Basel), 10, E12, doi: https://doi.org/10.3390/genes10010012.
Zhang, M., Fritsche, J., Roszik, J., Williams, L. J., Peng, X., Chiu, Y., Tsou, C.-C., Hoffgaard, F., Goldfinger, V., Schoor, O., Talukder, A., Forget, M. A., Haymaker, C., Bernatchez, C., Han, L., Tsang, Y.-H., Kong, K., Xu, X., Scott, K. L., Singh-Jasuja, H., Lizee, G., Liang, H., Weinschenk, T., Mills, G. B., and Hwu, P. (2018) RNA editing derived epitopes function as cancer antigens to elicit immune responses, Nat. Commun., 9, 3919, doi: https://doi.org/10.1038/s41467-018-06405-9.
Higuchi, M., Single, F. N., Kohler, M., Sommer, B., Sprengel, R., and Seeburg, P. H. (1993) RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency, Cell, 75, 1361–1370, doi: https://doi.org/10.1016/0092-8674(93)90622-W.
Egebjerg, J., Kukekov, V., and Heinemann, S. F. (1994) Intron sequence directs RNA editing of the glutamate receptor subunit GluR2 coding sequence, Proc. Natl. Acad. Sci. USA, 91, 10270–10274, doi: https://doi.org/10.1073/pnas.91.22.10270.
Grice, L. F., and Degnan, B. M. (2015) The origin of the ADAR gene family and animal RNA editing, BMC Evol. Biol., 15, 4, doi: https://doi.org/10.1186/s12862-015-0279-3.
Yablonovitch, A. L., Deng, P., Jacobson, D., and Li, J. B. (2017) The evolution and adaptation of A-to-I RNA editing, PLoS Genet., 13, e1007064, doi: https://doi.org/10.1371/journal.pgen.1007064.
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans, Nature, 391, 806–811, doi: https://doi.org/10.1038/35888.
Walkley, C. R., and Li, J. B. (2017) Rewriting the transcriptome: adenosine-to-inosine RNA editing by ADARs, Genome Biol., 18, 205, doi: https://doi.org/10.1186/s13059-017-1347-3.
Liscovitch-Brauer, N., Alon, S., Porath, H. T., Elstein, B., Unger, R., Ziv, T., Admon, A., Levanon, E. Y., Rosenthal, J. J. C., and Eisenberg, E. (2017) Trade-off between transcriptome plasticity and genome evolution in cephalopods, Cell, 169, 191–202.e11, doi: https://doi.org/10.1016/j.cell.2017.03.025.
Garrett, S., and Rosenthal, J. J. C. (2012) RNA editing underlies temperature adaptation in K+ channels from polar octopuses, Science, 335, 848–851, doi: https://doi.org/10.1126/science.1212795.
Iwamoto, K., Bundo, M., and Kato, T. (2009) Serotonin receptor 2C and mental disorders: genetic, expression, and RNA editing studies, RNA Biol., 6, 248–53, doi: https://doi.org/10.4161/rna.6.3.8370.
Barbon, A., and Barlati, S. (2011) Glutamate receptor RNA editing in health and disease, Biochemistry (Moscow), 76, 882–889, doi: https://doi.org/10.1134/S0006297911080037..
Gallo, A., Vukic, D., Michalik, D., O’Connell, M. A., and Keegan, L. P. (2017) ADAR RNA editing in human disease; more to it than meets the I, Hum. Genet., 136, 1265–1278, doi: https://doi.org/10.1007/s00439-017-1837-0.
Higuchi, M., Maas, S., Single, F. N., Hartner, J., Rozov, A., Burnashev, N., Feldmeyer, D., Sprengel, R., and Seeburg, P. H. (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2, Nature, 406, 78–81, doi: https://doi.org/10.1038/35017558.
Kawahara, Y., Ito, K., Sun, H., Aizawa, H., Kanazawa, I., and Kwak, S. (2004) RNA editing and death of motor neurons, Nature, 427, 801, doi: https://doi.org/10.1038/427801a.
Kwak, S., and Kawahara, Y. (2005) Deficient RNA editing of GluR2 and neuronal death in amyotropic lateral sclerosis, J. Mol. Med., 83, 110–120, doi: https://doi.org/10.1007/s00109-004-0599-z.
Lyddon, R., Dwork, A. J., Keddache, M., Siever, L. J., and Dracheva, S. (2013) Serotonin 2c receptor RNA editing in major depression and suicide, World J. Biol. Psychiatry, 14, 590–601, doi: https://doi.org/10.3109/15622975.2011.630406.
Sommer, B., Kohler, M., Sprengel, R., and Seeburg, P. H. (1991) RNA editing in brain controls a determinant of ion flow in glutamate-gated channels, Cell, 67, 11–19, doi: https://doi.org/10.1016/0092-8674(91)90568-J.
Patterson, J. B., and Samuel, C. E. (1995) Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase, Mol. Cell. Biol., 15, 5376–5388.
Wright, A., and Vissel, B. (2012) The essential role of AMPA receptor GluR2 subunit RNA editing in the normal and diseased brain, Front. Mol. Neurosci., 5, 34, doi: https://doi.org/10.3389/fnmol.2012.00034.
Funding
Funding. This work is supported by the Russian Science Foundation (project no. 17-15-01229).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest. The authors declare no conflict of interest in financial of any other sphere.
Ethical approval. This paper contains no studies using animal or human subjects.
Additional information
Published in Russian in Biokhimiya, 2019, Vol. 84, No. 8, pp. 1129–1138.
Rights and permissions
About this article
Cite this article
Goncharov, A.O., Kliuchnikova, A.A., Nasaev, S.S. et al. RNA Editing by ADAR Adenosine Deaminases: From Molecular Plasticity of Neural Proteins to the Mechanisms of Human Cancer. Biochemistry Moscow 84, 896–904 (2019). https://doi.org/10.1134/S0006297919080054
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297919080054