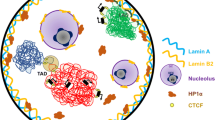
Abstract
Chromosomes are intricately folded and packaged in the cell nucleus and interact with the nuclear envelope. This complex nuclear architecture has a profound effect on how the genome works and how the cells function. The main goal of review is to highlight recent studies on the effect of chromosome–nuclear envelope interactions on chromatin folding and function in the nucleus. The data obtained suggest that chromosome–nuclear envelope attachments are important for the organization of nuclear architecture in various organisms. A combination of experimental cell biology methods with computational modeling offers a unique opportunity to explore the fundamental relationships between different aspects of 3D genome organization in greater details. This powerful interdisciplinary approach could reveal how the organization and function of the genome in the nuclear space is affected by the chromosome–nuclear envelope attachments and will enable the development of novel approaches to regulate gene expression.
Similar content being viewed by others
Abbreviations
- Chr:
-
chromosome
- DamID:
-
DNA adenine methyltransferase identification
- FISH:
-
fluorescence in situ hybridization
- Hi-C:
-
chromosome conformation capture
- LAD:
-
lamina-associated domain
- NE:
-
nuclear envelope
- TAD:
-
topologically associated domain
References
Lemaitre, C., and Bickmore, W. A. (2015) Chromatin at the nuclear periphery and the regulation of genome functions, Histochem. Cell Biol., 144, 111–122.
Ciabrelli, F., and Cavalli, G. (2015) Chromatin-driven behavior of topologically associating domains, J. Mol. Biol., 427, 608–625.
Barton, L. J., Soshnev, A. A., and Geyer, P. K. (2015) Networking in the nucleus: a spotlight on LEM-domain proteins, Curr. Opin. Cell Biol., 34, 1–8.
Nguyen, H. Q., and Bosco, G. (2015) Gene positioning effects on expression in eukaryotes, Annu. Rev. Genet., 49, 627–646.
Grosberg, A. Y., Nechaev, S. K., and Shakhnovich, E. I. (1988) The role of topological constraints in the kinetics of collapse of macromolecules, J. Phys. (Paris), 49, 2095–2100.
Lebedev, D. V., Filatov, M. V., Kuklin, A. I., Islamov, A. K., Kentzinger, E., Pantina, R., Toperverg, B. P., and Isaev-Ivanov, V. V. (2005) Fractal nature of chromatin organization in interphase chicken erythrocyte nuclei: DNA structure exhibits biphasic fractal properties, FEBS Lett., 579, 1465–1468.
Gursoy, G., Xu, Y., Kenter, A. L., and Liang, J. (2014) Spatial confinement is a major determinant of the folding landscape of human chromosomes, Nucleic Acids Res., 42, 8223–8230.
Mateos-Langerak, J., Bohn, M., de Leeuw, W., Giromus, O., Manders, E. M., Verschure, P. J., Indemans, M. H., Gierman, H. J., Heermann, D. W., van Driel, R., and Goetze, S. (2009) Spatially confined folding of chromatin in the interphase nucleus, Proc. Natl. Acad. Sci. USA, 106, 3812–3817.
Pickersgill, H., Kalverda, B., de Wit, E., Talhout, W., Fornerod, M., and van Steensel, B. (2006) Characterization of the Drosophila melanogaster genome at the nuclear lamina, Nat. Genet., 38, 1005–1014.
Guelen, L., Pagie, L., Brasset, E., Meuleman, W., Faza, M. B., Talhout, W., Eussen, B. H., de Klein, A., Wessels, L., de Laat, W., and van Steensel, B. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions, Nature, 453, 948–951.
Hochstrasser, M., Mathog, D., Gruenbaum, Y., Saumweber, H., and Sedat, J. W. (1986) Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster, J. Cell. Biol., 102, 112–123.
Gonzalez-Sandoval, A., and Gasser, S. M. (2016) On TADs and LADs: spatial control over gene expression, Trends Genet., 32, 485–495.
Van Bemmel, J. G., Pagie, L., Braunschweig, U., Brugman, W., Meuleman, W., Kerkhoven, R. M., and van Steensel, B. (2010) The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome, PLoS One, 5, e15013.
Shevelyov, Y. Y., Lavrov, S. A., Mikhaylova, L. M., Nurminsky, I. D., Kulathinal, R. J., Egorova, K. S., Rozovsky, Y. M., and Nurminsky, D. I. (2009) The B-type lamin is required for somatic repression of testis-specific gene clusters, Proc. Natl. Acad. Sci. USA, 106, 3282–3287.
Kohwi, M., Lupton, J. R., Lai, S. L., Miller, M. R., and Doe, C. Q. (2013) Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila, Cell, 152, 97–108.
Amendola, M., and van Steensel, B. (2015) Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells, EMBO Rep., 16, 610–617.
Hochstrasser, M., and Sedat, J. W. (1987) Three-dimensional organization of Drosophila melanogaster interphase nuclei. II. Chromosome spatial organization and gene regulation, J. Cell Biol., 104, 1471–1483.
Hochstrasser, M., and Sedat, J. W. (1987) Three-dimensional organization of Drosophila melanogaster interphase nuclei. I. Tissue-specific aspects of polytene nuclear architecture, J. Cell Biol., 104, 1455–1470.
Chen, H., Zheng, X., and Zheng, Y. (2014) Age-associated loss of lamin-B leads to systemic inflammation and gut hyperplasia, Cell, 159, 829–843.
Cremer, T., and Cremer, M. (2010) Chromosome territories, Cold Spring Harb. Perspect. Biol., 2, a003889.
Cremer, T., and Cremer, C. (2001) Chromosome territories, nuclear architecture and gene regulation in mammalian cells, Nat. Rev. Genet., 2, 292–301.
Bauer, C. R., Hartl, T. A., and Bosco, G. (2012) Condensin II promotes the formation of chromosome territories by inducing axial compaction of polyploid interphase chromosomes, PLoS Genet., 8, e1002873.
Mathog, D., Hochstrasser, M., Gruenbaum, Y., Saumweber, H., and Sedat, J. (1984) Characteristic folding pattern of polytene chromosomes in Drosophila salivary gland nuclei, Nature, 308, 414–421.
Lieberman-Aiden, E., van Berkum, N. L., Williams, L., Imakaev, M., Ragoczy, T., Telling, A., Amit, I., Lajoie, B. R., Sabo, P. J., Dorschner, M. O., Sandstrom, R., Bernstein, B., Bender, M. A., Groudine, M., Gnirke, A., Stamatoyannopoulos, J., Mirny, L. A., Lander, E. S., and Dekker, J. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome, Science, 326, 289–293.
Sexton, T., Yaffe, E., Kenigsberg, E., Bantignies, F., Leblanc, B., Hoichman, M., Parrinello, H., Tanay, A., and Cavalli, G. (2012) Three-dimensional folding and functional organization principles of the Drosophila genome, Cell, 148, 458–472.
Ulianov, S. V., Khrameeva, E. E., Gavrilov, A. A., Flyamer, I. M., Kos, P., Mikhaleva, E. A., Penin, A. A., Logacheva, M. D., Imakaev, M. V., Chertovich, A., Gelfand, M. S., Shevelyov, Y. Y., and Razin, S. V. (2016) Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains, Genome Res., 26, 70–84.
Battulin, N., Fishman, V. S., Mazur, A. M., Pomaznoy, M., Khabarova, A. A., Afonnikov, D. A., Prokhortchouk, E. B., and Serov, O. L. (2015) Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach, Genome Biol., 16, 77.
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., Hu, M., Liu, J. S., and Ren, B. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions, Nature, 485, 376–380.
Ong, C. T., and Corces, V. G. (2014) CTCF: an architectural protein bridging genome topology and function, Nat. Rev. Genet., 15, 234–246.
Cmarko, D., Verschure, P. J., Martin, T. E., Dahmus, M. E., Krause, S., Fu, X. D., van Driel, R., and Fakan, S. (1999) Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection, Mol. Biol. Cell, 10, 211–223.
Eagen, K. P., Hartl, T. A., and Kornberg, R. D. (2015) Stable chromosome condensation revealed by chromosome conformation capture, Cell, 163, 934–946.
Filion, G. J., van Bemmel, J. G., Braunschweig, U., Talhout, W., Kind, J., Ward, L. D., Brugman, W., de Castro, I. J., Kerkhoven, R. M., Bussemaker, H. J., and van Steensel, B. (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells, Cell, 143, 212–224.
Kharchenko, P. V., Alekseyenko, A. A., Schwartz, Y. B., Minoda, A., Riddle, N. C., Ernst, J., Sabo, P. J., Larschan, E., Gorchakov, A. A., Gu, T., Linder-Basso, D., Plachetka, A., Shanover, G., Tolstorukov, M. Y., Luquette, L. J., Xi, R., Jung, Y. L., Park, R. W., Bishop, E. P., Canfield, T. K., Sandstrom, R., Thurman, R. E., MacAlpine, D. M., Stamatoyannopoulos, J. A., Kellis, M., Elgin, S. C., Kuroda, M. I., Pirrotta, V., Karpen, G. H., and Park, P. J. (2011) Comprehensive analysis of the chromatin landscape in Drosophila melanogaster, Nature, 471, 480–485.
Osborne, C. S., Chakalova, L., Brown, K. E., Carter, D., Horton, A., Debrand, E., Goyenechea, B., Mitchell, J. A., Lopes, S., Reik, W., and Fraser, P. (2004) Active genes dynamically colocalize to shared sites of ongoing transcription, Nat. Genet., 36, 1065–1071.
Cheutin, T., and Cavalli, G. (2012) Progressive polycomb assembly on H3K27me3 compartments generates polycomb bodies with developmentally regulated motion, PLoS Genet., 8, e1002465.
Li, H. B., Ohno, K., Gui, H., and Pirrotta, V. (2013) Insulators target active genes to transcription factories and polycomb-repressed genes to polycomb bodies, PLoS Genet., 9, e1003436.
Finlan, L. E., Sproul, D., Thomson, I., Boyle, S., Kerr, E., Perry, P., Ylstra, B., Chubb, J. R., and Bickmore, W. A. (2008) Recruitment to the nuclear periphery can alter expression of genes in human cells, PLoS Genet., 4, e1000039.
Reddy, K. L., Zullo, J. M., Bertolino, E., and Singh, H. (2008) Transcriptional repression mediated by repositioning of genes to the nuclear lamina, Nature, 452, 243–247.
Henikoff, S., and Dreesen, T. D. (1989) Trans-inactivation of the Drosophila brown gene: evidence for transcriptional repression and somatic pairing dependence, Proc. Natl. Acad. Sci. USA, 86, 6704–6708.
Tolhuis, B., Blom, M., Kerkhoven, R. M., Pagie, L., Teunissen, H., Nieuwland, M., Simonis, M., de Laat, W., van Lohuizen, M., and van Steensel, B. (2011) Interactions among Polycomb domains are guided by chromosome architecture, PLoS Genet., 7, e1001343.
Bantignies, F., Roure, V., Comet, I., Leblanc, B., Schuettengruber, B., Bonnet, J., Tixier, V., Mas, A., and Cavalli, G. (2011) Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila, Cell, 144, 214–226.
Denholtz, M., Bonora, G., Chronis, C., Splinter, E., de Laat, W., Ernst, J., Pellegrini, M., and Plath, K. (2013) Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization, Cell Stem Cell, 13, 602–616.
Papantonis, A., Kohro, T., Baboo, S., Larkin, J. D., Deng, B., Short, P., Tsutsumi, S., Taylor, S., Kanki, Y., Kobayashi, M., Li, G., Poh, H. M., Ruan, X., Aburatani, H., Ruan, Y., Kodama, T., Wada, Y., and Cook, P. R. (2012) TNFalpha signals through specialized factories where responsive coding and miRNA genes are transcribed, EMBO J., 31, 4404–4414.
Schoenfelder, S., Sexton, T., Chakalova, L., Cope, N. F., Horton, A., Andrews, S., Kurukuti, S., Mitchell, J. A., Umlauf, D., Dimitrova, D. S., Eskiw, C. H., Luo, Y., Wei, C. L., Ruan, Y., Bieker, J. J., and Fraser, P. (2010) Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells, Nat. Genet., 42, 53–61.
Kind, J., Pagie, L., Ortabozkoyun, H., Boyle, S., de Vries, S. S., Janssen, H., Amendola, M., Nolen, L. D., Bickmore, W. A., and van Steensel, B. (2013) Single-cell dynamics of genome–nuclear lamina interactions, Cell, 153, 178–192.
Kinney, N. A., Onufriev, A. V., and Sharakhov, I. V. (2015) Quantified effects of chromosome–nuclear envelope attachments on 3D organization of chromosomes, Nucleus, 6, 212–224.
Chubb, J. R., Boyle, S., Perry, P., and Bickmore, W. A. (2002) Chromatin motion is constrained by association with nuclear compartments in human cells, Curr. Biol., 12, 439–445.
Hubner, M., and Spector, D. (2010) Chromatin dynamics, Annu. Rev. Biophys., 39, 471–489.
Agmon, N., Liefshitz, B., Zimmer, C., Fabre, E., and Kupiec, M. (2013) Effect of nuclear architecture on the efficiency of double-strand break repair, Nat. Cell Biol., 15, 694–699.
Sexton, T., and Cavalli, G. (2015) The role of chromosome domains in shaping the functional genome, Cell, 160, 1049–1059.
Fanucchi, S., Shibayama, Y., Burd, S., Weinberg, M. S., and Mhlanga, M. M. (2013) Chromosomal contact permits transcription between coregulated genes, Cell, 155, 606–620.
Hartl, T. A., Smith, H. F., and Bosco, G. (2008) Chromosome alignment and transvection are antagonized by condensin II, Science, 322, 1384–1387.
Zhimulev, I. F. (1996) Morphology and structure of polytene chromosomes, in Advances in Genetics (Hall, J. C., ed.) Vol. 34, Academic Press, San Diego, pp. 1–490.
Demakov, S. A., Vatolina, T. Yu., Babenko, V. N., Semeshin, V. F., Belyaeva, E. S., and Zhimulev, I. F. (2011) Protein composition of interband regions in polytene and cell line chromosomes of Drosophila melanogaster, BMC Genom., 12, 566.
Belyaeva, E. S., Goncharov, F. P., Demakova, O. V., Kolesnikova, T. D., Boldyreva, L. V., Semeshin, V. F., and Zhimulev, I. F. (2012) Late replication domains in polytene and non-polytene cells of Drosophila melanogaster, PLoS One, 7, e30035.
Vatolina, T. Y., Boldyreva, L. V., Demakova, O. V., Demakov, S. A., Kokoza, E. B., Semeshin, V. F., Babenko, V. N., Goncharov, F. P., Belyaeva, E. S., and Zhimulev, I. F. (2011) Identical functional organization of nonpolytene and polytene chromosomes in Drosophila melanogaster, PLoS One, 6, e25960.
Zhimulev, I. F., Zykova, T. Y., Goncharov, F. P., Khoroshko, V. A., Demakova, O. V., Semeshin, V. F., Pokholkova, G. V., Boldyreva, L. V., Demidova, D. S., Babenko, V. N., Demakov, S. A., and Belyaeva, E. S. (2014) Genetic organization of interphase chromosome bands and interbands in Drosophila melanogaster, PLoS One, 9, e101631.
Mathog, D., and Sedat, J. W. (1989) The three-dimensional organization of polytene nuclei in male Drosophila melanogaster with compound XY or ring X chromosomes, Genetics, 121, 293–311.
Sharakhov, I. V., Sharakhova, M. V., Mbogo, C. M., Koekemoer, L. L., and Yan, G. (2001) Linear and spatial organization of polytene chromosomes of the African malaria mosquito Anopheles funestus, Genetics, 159, 211–218.
Sharakhova, M. V., George, P., Brusentsova, I. V., Leman, S. C., Bailey, J. A., Smith, C. D., and Sharakhov, I. V. (2010) Genome mapping and characterization of the Anopheles gambiae heterochromatin, BMC Genom., 11, 459.
Beliveau, B. J., Joyce, E. F., Apostolopoulos, N., Yilmaz, F., Fonseka, C. Y., McCole, R. B., Chang, Y., Li, J. B., Senaratne, T. N., Williams, B. R., Rouillard, J. M., and Wu, C. T. (2012) Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes, Proc. Natl. Acad. Sci. USA, 109, 21301–21306.
Cremer, M., Kupper, K., Wagler, B., Wizelman, L., von Hase, J., Weiland, Y., Kreja, L., Diebold, J., Speicher, M. R., and Cremer, T. (2003) Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei, J. Cell Biol., 162, 809–820.
Artemov, G., Bondarenko, S., Sapunov, G., and Stegniy, V. (2015) Tissue-specific differences in the spatial interposition of X-chromosome and 3R chromosome regions in the malaria mosquito Anopheles messeae Fall, PLoS One, 10, e0115281.
Stegnii, V. N. (1987) Systemic reorganization of the architectonics of polytene chromosomes in the onto-and phylogenesis of malaria mosquitoes, Genetika, 23, 821–827.
Bondarenko, S. M., Artemov, G. N., Sharakhov, I. V., and Stegniy, V. N. (2017) Tissue-specific features of the X chromosome and nucleolus spatial dynamics in a malaria mosquito, Anopheles atroparvus, PLoS One, 12, e0171290.
Stegnii, V. N. (1979) Reorganization of the structure of interphase nuclei during the onto-and phylogeny of malaria mosquitoes, Dokl. Akad. Nauk. SSSR, 249, 1231–1234.
Stegnii, V. N., and Sharakhova, M. V. (1991) Systemic reorganization of the architechtonics of polytene chromosomes in onto-and phylogenesis of malaria mosquitoes. Structural features regional of chromosomal adhesion to the nuclear membrane, Genetika, 27, 828–835.
Ollion, J., Cochennec, J., Loll, F., Escude, C., and Boudier, T. (2013) TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization, Bioinformatics, 29, 1840–1841.
Stegnii, V. N. (1987) Systemic reorganization of the archi-tectonics of polytene chromosomes in the onto-and phylogenesis of malarial mosquitoes. II. Species specificity in the pattern of chromosome relations with the nuclear envelope of nutrient ovarian cells, Genetika, 23, 1194–1199.
Pombi, M., Caputo, B., Simard, F., Di Deco, M. A., Coluzzi, M., della Torre, A., Costantini, C., Besansky, N. J., and Petrarca, V. (2008) Chromosomal plasticity and evolutionary potential in the malaria vector Anopheles gambiae sensu stricto: insights from three decades of rare paracentric inversions, BMC Evol. Biol., 8, 309.
Pevzner, P., and Tesler, G. (2003) Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution, Proc. Natl. Acad. Sci. USA, 100, 7672–7677.
Alekseyev, M. A. (2008) Multi-break rearrangements and breakpoint re-uses: from circular to linear genomes, J. Comput. Biol., 15, 1117–1131.
Gandhi, M., Evdokimova, V., and Nikiforov, Y. E. (2010) Mechanisms of chromosomal rearrangements in solid tumors: the model of papillary thyroid carcinoma, Mol. Cell Endocrinol., 321, 36–43.
Marshall, W. F. (2002) Order and disorder in the nucleus, Curr. Biol., 12, R185–192.
Folle, G. A. (2008) Nuclear architecture, chromosome domains and genetic damage, Mutat. Res., 658, 172–183.
Fudenberg, G., Getz, G., Meyerson, M., and Mirny, L. A. (2011) High order chromatin architecture shapes the landscape of chromosomal alterations in cancer, Nat. Biotechnol., 29, 1109–1113.
Gandhi, M., Medvedovic, M., Stringer, J. R., and Nikiforov, Y. E. (2006) Interphase chromosome folding determines spatial proximity of genes participating in carcinogenic RET/PTC rearrangements, Oncogene, 25, 2360–2366.
Duan, Z., Andronescu, M., Schutz, K., McIlwain, S., Kim, Y. J., Lee, C., Shendure, J., Fields, S., Blau, C. A., and Noble, W. S. (2010) A three-dimensional model of the yeast genome, Nature, 465, 363–367.
Khrameeva, E. E., Fudenberg, G., Gelfand, M. S., and Mirny, L. A. (2016) History of chromosome rearrangements reflects the spatial organization of yeast chromosomes, J. Bioinform. Comput. Biol., 14, 1641002.
Cheutin, T., Bantignies, F., Leblanc, B., and Cavalli, G. (2010) Chromatin folding: from linear chromosomes to the 4D nucleus, Cold Spring Harb. Symp. Quant. Biol., 75, 461–473.
Peric-Hupkes, D., and van Steensel, B. (2010) Role of the nuclear lamina in genome organization and gene expression, Cold Spring Harb. Symp. Quant. Biol., 75, 517–524.
Nagano, T., Lubling, Y., Stevens, T. J., Schoenfelder, S., Yaffe, E., Dean, W., Laue, E. D., Tanay, A., and Fraser, P. (2013) Single-cell Hi-C reveals cell-to-cell variability in chromosome structure, Nature, 502, 59–64.
Mirny, L. A. (2011) The fractal globule as a model of chromatin architecture in the cell, Chromosome Res., 19, 37–51.
Zhang, B., and Wolynes, P. G. (2015) Topology, structures, and energy landscapes of human chromosomes, Proc. Natl. Acad. Sci. USA, 112, 6062–6067.
Li, Q. J., Tjong, H., Li, X., Gong, K., Zhou, X. J., Chiolo, I., and Alber, F. (2017) The three-dimensional genome organization of Drosophila melanogaster through data integration, Genome Biol., 18, 145.
Cook, P. R., and Marenduzzo, D. (2009) Entropic organization of interphase chromosomes, J. Cell Biol., 186, 825–834.
Mirny, L. (2011) The fractal globule as a model of chromatin architecture in the cell, Chromosome Res., 19, 37–51.
Wong, H., Marie-Nelly, H., Herbert, S., Carrivain, P., Blanc, H., Koszul, R., Fabre, E., and Zimmer, C. (2012) A predictive computational model of the dynamic 3D interphase yeast nucleus, Curr. Biol., 22, 1881–1890.
Wong, H., Arbona, J. M., and Zimmer, C. (2013) How to build a yeast nucleus, Nucleus, 4, 361–366
Rowley, M. J., and Corces, V. G. (2016) The three-dimensional genome: principles and roles of long-distance interactions, Curr. Opin. Cell Biol., 40, 8–14.
Kinney, N., Sharakhov, I. V., and Onufriev, A. V. (2014) Investigation of the chromosome regions with significant affinity for the nuclear envelope in fruit fly–a model-based approach, PLoS One, 9, e91943.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sharakhov, I.V., Bondarenko, S.M., Artemov, G.N. et al. The Role of Chromosome–Nuclear Envelope Attachments in 3D Genome Organization. Biochemistry Moscow 83, 350–358 (2018). https://doi.org/10.1134/S0006297918040065
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297918040065