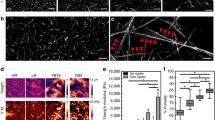
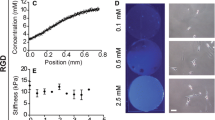
Abstract
Adhesion-mediated migration is required in a number of physiological and pathological processes. A further quantitative understanding of the relationship between cell migration and cell-substratum adhesiveness may aid in therapeutic or tissue engineering applications. The aim of this work was to quantify three-dimensional cell migration as a function of increasing cell-substratum adhesiveness within reconstituted collagen gels. Cell-substratum adhesiveness was controlled by grafting additional adhesive peptides containing the well-characterized arginine-glycine-aspartic acid sequence to collagen. The three-dimensional migration of multiple individual cells was tracked in real time in an automated fashion for extended periods. Cell displacements were statistically analyzed and fit to a correlated persistent random walk model to estimate root-mean-square speed, directional persistence time, and random motility coefficient. Based on model parameter estimates, cell speed was found to be a monotonically decreasing function of increasing substratum adhesiveness, while the directional persistence time and random motility coefficient exhibited a biphasic dependence, with maximum values at approximately intermediate concentrations of grafted adhesive peptide and hence intermediate cell-substratum adhesiveness. In conclusion, these studies suggest an optimal adhesiveness for three-dimensional random migration, consistent with previous studies on two-dimensional surfaces. However, the maximum in random motility corresponded to a maximum in directional persistence, not in cell speed. © 2000 Biomedical Engineering Society.
PAC00: 8780Rb, 8714Ee, 8717Jj, 8715La, 8270Gg
Similar content being viewed by others
REFERENCES
1Akiyama, S. K.Integrins in cell adhesion and signaling. Hum Cell.9:181-6, 1996.
2Albelda, S. M., S. A. Mette, D. E. Elder, R. Stewart, L. Damjanovich, M. Herlyn, and C. A. Buck. Integrin distribution in malignant melanoma: association of the beta 3 subunit with tumor progression. Cancer Res.50:6757-64, 1990.
3Alt, W. Correlation Analysis of Two-Dimensional Locomotion Paths, Biological Motion, edited by W. Alt and G. Hoffmann. Berlin: Springer, 1990, pp.254-268.
4Aznavoorian, S., M. L. Stracke, J. Parsons, J. McClanahan, and L. A. Liotta. Integrin alphavbeta3 mediates chemotactic and haptotactic motiliby in human melanoma cells through different signaling pathways. J. Biol. Chem.271:3247-54, 1996.
5Bergman, A., and K. Zygourakis. Lymphocyte Motility and Adhesion on Fibronectin Surfaces Through the Beta-1 Integrin. 1998 AICHE Annual Meeting. Paper 278a, 1998 (unpublished).
6Berman, A., G. Morozevich, I. Karmansky, A. Gleiberman, and V. Bychkova. Adhesion of mouse heptocytes to type I collagen: role of supramolecular forms and effect of proteolytic degradation. Biochem. Biophys. Res. Commun.194:351-7, 1993.
7Dedhar, S., E. Ruoslahti, and M. D. Pierschbacher. A cell surface receptor complex for collagen type I recognizes the Arg-Gly-Asp sequence. J. Cell Biol.104:585-93, 1987.
8Deryugina, E. I., M. A. Bourdon, R. A. Reisfeld, and A. Strongin. Remodeling of collagen matrix by human tumor cells requires activation and cell surface association of matrix metalloproteinase-2. Cancer Res.58:3743-50, 1998.
9Dickinson, R. B., S. Guido, and R. T. Tranquillo. Biased cell migration of fibroblasts exhibiting contact guidance in oriented collagen gels. Ann. Biomed. Eng.22:342-56, 1994.
10Dickinson, R. B., and R. T. Tranquillo. Optimal estimation of cell movement indices from the statistical analysis of cell tracking data. AIChE. J.39:1995-2010, 1993.
11DiMilla, P. A., J. A. Stone, J. A. Quinn, S. M. Albelda, and D. A. Lauffenburger. Maximal migration of human smooth muscle cells on fibronectin and type IV collagen occurs at an intermediate attachment strength. J. Cell Biol.122:729-37, 1993.
12Duband, J. L., S. Dufour, S. S. Yamada, K. M. Yamada, and J. P. Thiery. Neural crest cell locomotion induced by antibodies to beta 1 integrins. A tool for studying the roles of substratum molecular avidity and density in migration. J. Cell. Sci.98:517-32, 1991.
13Dunn, G. A.Characterizing a kinesis response: time averaged measures of cell speed and directional persistence. Agents Actions Suppl.12:14-33, 1983.
14Friedl, P., E. B. Brocker, and K. S. Zanker. Integrins, cell matrix interactions, and cell migration strategies: fundamental differences in leukocytes and tumor cells. Cell Adhes Commun.6:225-36, 1998.
15Friedl, P., F. Entschladen, C. Conrad, B. Niggemann, and K. S. Zanker. CD4+T lymphocytes migrating in three-dimensional collagen lattices lack focal adhesions and utilize beta1 integrin-independent strategies for polarization, interaction with collagen fibers and locomoation. Eur. J. Immunol.28:2331-43, 1998.
16Friedl, P., K. Maaser, C. E. Klein, B. Niggemann, G. Krohne, and K. S. Zanker. Migration of highly aggressive MV3 melanoma cells in 3-dimensional collagen lattices results in local matrix reorganization and shedding of alpha2 and beta1 integrins and CD44. Cancer Res.57:2061-70, 1997.
17Friedl, P., K. S. Zanker, and E. B. Brocker. Cell migration strategies in 3-D extracellular matrix: differences in morphology, cell matrix interactions, and integrin function [In Process Citation]. Microsc. Res. Tech.43:369-78, 1998.
18Grinnell, F.Migration of human neutrophils in hydrated collagen lattices. J. Cell. Sci.58:95-108, 1982.
19Gunzer, M., E. Kampgen, E. B. Brocker, K. S. Zanker, and P. Friedl. Migration of dendritic cells in 3D-collagen lattices. Visualisation of dynamic interactions with the substratum and the distribution of surface structures via a novel confocal reflection imaging technique. Adv. Exp. Med. Biol.417:97-103, 1997.
20Haas, T., S. J. Davis, and J. A. Madri. Three-dimensional type I collagen lattices induce coordinate expression of matrix metalloproteinases MT1-MMP and MMP-2 in microvascular endothelial cells. J. Biol. Chem.273:3604-10, 1998.
21Huttenlocher, A., M. H. Ginsberg, and A. F. Horwitz. Modulation of cell migration by integrin-mediated cytoskeletal linkages and ligand-binding affinity. J. Cell Biol.134:1551-62, 1996.
22Huttenlocher, A., R. R. Sandborg, and A. F. Horwitz. Adhesion in cell migration. Curr. Opin. Cell Biol.7:697-706, 1995.
23Keely, P. J., A. M. Fong, M. M. Zutter, and S. A. Santoro. Alteration of collagen-dependent adhesion, motility, and morphogenesis by the expression of antisense alpha 2 integrin mRNA in mammary cells. J. Cell. Sci.108:595-607, 1995.
24Kramer, R. H., M. Vu, Y. F. Cheng, and D. M. Ramos. Iintegrin expression in malignant melanoma. Cancer Metastasis Rev.10:49-59, 1991.
25Kramer, R. H., and N. Marks. Identification of integrin collagen receptors on human melanoma cells. J. Biol. Chem.264:4684-8, 1989.
26Kuntz, R. M., and W. M. Saltzman. Neutrophil motility in extracellular matrix gels: mesh size and adhesion affect speed of migration. Biophys. J.72:1472-80, 1997.
27Lauffenburger, D. A., and A. F. Horwitz. Cell migration: a physically integrated molecular process. Cell84:359-69, 1996.
28Morla, A., Z. Zhang, and E. Ruoslahti. Superfibronectin is a functionally distinct form of fibronectin. Nature (London)367:193-6, 1994.
29Myles, J. L., B. T. Burgess, and R. B. Dickinson. Modification of the Adhesive Properties of Collagen by Covalent Grafting with RGD Peptides. (in press).
30Palecek, S. P., A. Huttenlocher, A. F. Horwitz, and D. A. Lauffenburger. Physical and biochemical regulation of integrin release during rear detachment of migrating cells. J. Cell. Sci.111:929-40, 1998.
31Palecek, S. P., J. C. Loftus, M. H. Ginsberg, D. A. Lauffenburger, and A. F. Horwitz. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness [published erratum appears in Nature 1997 Jul 10;388(6638):210]Nature (London)385:537-40, 1997.
32Pierschbacher, M. D., E. G. Hayman, and E. Ruoslahti. The cell attachment determinant in fibronectin. J. Cell. Biochem.28:115-26, 1985.
33Pierschbacher, M. D., and E. Ruoslahti. Variants of the cell recognition site of fibronectin that retain attachment-promoting activity. Proc. Natl. Acad. Sci. USA81:5985-8, 1984.
34Ruoslahti, E.RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol.12:697-715, 1996.
35Ruoslahti, E., S. Suzuki, E. G. Hayman, C. R. Ill, and M. D. Pierschbacher. Purification and characterization of vitronectin. Methods Enzymol.144:430-7, 1987.
36Sheetz, M. P.Cell migration by graded attachment to substrates and contraction. Semin. Cell Biol.5:149-55, 1994.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Burgess, B.T., Myles, J.L. & Dickinson, R.B. Quantitative Analysis of Adhesion-Mediated Cell Migration in Three-Dimensional Gels of RGD-Grafted Collagen. Annals of Biomedical Engineering 28, 110–118 (2000). https://doi.org/10.1114/1.259
Issue Date:
DOI: https://doi.org/10.1114/1.259