Abstract
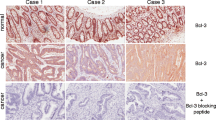
The BCR gene is located on human chromosome 22.The normal cellular BCR gene encodes a 160,000-daltonphosphoprotein associated with a serine/threonine kinaseactivity. The BCR protein is involved in signal transduction. We investigated the expression ofthe BCR protein in hepatocellular carcinoma (HCC),surrounding noncancerous liver tissue, liver cirrhosis(LC), chronic hepatitis (CH), and normal liver with immunohistochemistry and a western blotanalysis. BCR immunoreactivity was detected using amonoclonal antibody. In normal liver, and both CH and LCwithout association of HCC, the immunoreactivity of the BCR protein was minimal. In contrast, 73% (22of 30) of noncancerous liver tissue adjacent to the HCCand 40% (12 of 30) of HCC expressed BCR protein; thisdifference was statistically significant (P < 0.01). The expression of the BCR protein expressioncorrelated with the degree of histologicaldifferentiation of HCC (P < 0.05). In addition, theamplification of BCR protein in noncancerous cells wassupported by the detection of specific protein using awestern blot analysis. In two cases, the expression ofBCR protein occurred only in overtly malignant HCCcells. As a result, the expression of the BCR protein may be associated with oncogenesis in humanHCC.
Similar content being viewed by others
REFERENCES
Statics and Information Department, Minister's Secretariat, Ministry and Welfare: Mortality Statistics from Malignant Neoplasms, 1972–1984: Special Report of Viral Statistics in Japan. Tokyo, Ministry of Health and Welfare, 1986, pp 48-73
Tanaka K, Hirohata T, Koga S, Sugimachi K, Kanematu T, Ohryohji F, Nawata H, et al: Hepatitis C and hepatitis B in the etiology of hepatocellular carcinoma in the Japanese population. Cancer Res 51:2842-2847, 1991
Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y: Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA 87:6547-6549, 1990
Liver Cancer Study Group of Japan: Primary Liver Cancer in Japan: Sixth Report. Cancer 60:1400-1411, 1987
Tsuda H, Hirohashi S, Shimosato Y, Terada M, Hasegawa H: Clonal origin of atypical adenomatous hyperplasia of the liver and clonal identity with hepatocellular carcinoma. Gastroenterology 95:1664-1665, 1988
Sugihara S, Nakashima O, Kiyomatsu K, Edamitsu O, Kojiro M: Pathological study on hyperplastic nodule of the liver: A special reference to the containment of cancerous foci. Acta Hepatol Jpn 68:324-330, 1990 (in Japanese)
Rowley JD: A new consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and Giemsa staining. Nature 243:290-293, 1973
Groffen J, Stephenson JR, Heisterkamp M, de Klein A, Bartram CR, Grosueld G: Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell 36:93-99, 1984
Shtivelman E, Lifschitz B, Gale RP, Canaani E: Fused transcript of abl and bcr genes in chronic myelogenous Leukemia. Nature 315:550-554, 1985
Hermans A, Heisterkamp N, von Lindern M, Van Boal S, Meijen D, van der Plas D, Wiedemann LM, Groffen J, Bootsma D, Grosveld G: Unique fusion of bcr and c-abl genes in Philadelphia chromosome positive acute lymphoblastic leukemia. Cell 51:33-40, 1987
Maru Y, Witte ON: The BCR gene encodes a novel serine/threonine kinase activity within a single exon. Cell 67:459-468, 1991
Heisterkamp N, Stam K, Groffen J, de Klein A, and Grosveld G: Structural organization of the bcr gene and its role in the Ph1 translocation. Nature 315:758-761, 1985
Hariharan IK, Adams JM: cDNA sequence for human bcr, the gene that translocates to the abl oncogene in chronic myeloid leukemia. EMBO J 6:115-119, 1987
Collins S, Colman H, Groudine M: Expression of bcr and bcr-abl fusion transcripts in normal and leukemic cells. Mol Cell Biol 7:2870-2876, 1987
Stam K, Heisterkamp N, Reynolds FH Jr, Groffen J: Evidence that the ph1 gene encodes a 160,000-dalton phosphoprotein with associated kinase activity. Mol Cell Biol 7:1955-1960, 1987
Li W, Dreazen O, Kloetzer W, Gale RP, Arlinghaus RB: Characterization of bcr gene products in hematopoietic cells. Oncogene 4:127-138, 1988
Timmons MS, Whitte ON: Structural characterization of the BCR gene product. Oncogene 4:559-567, 1989
Hall C, Monfries C, Smith P, Lim HH, Ahmed RKS, Vanniasingham V, Leung T, Lim L: Novel human brain cDNA encoding a 34,000 Mr protein n-chimaerin, related to both the regulatory domain of protein kinase C and BCR, the product of the breakpoint cluster region gene. J Mol Biol 211:11-16, 1990
Diekmann D, Brill S, Garrett MD, Totty N, Hsuan J, Monfries C, Hall C, Lim L, Hall A: BCR encodes a GTPase activating protein for p21rac. Nature 351:400-402, 1991
Shou C, Fransworth CL, Neel BG, Feig LA: Molecular cloning of cDNAs encoding a guanine-nucleotid-releasing factor for Ras p21. Nature 358:351-354, 1992
Cantley L, Auger K, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S: Oncogenes and signal transduction. Cell 64:281-302, 1991
Stam K, Heisterkamp N, Grosveld G, deKlein A, Verma RS, Coleman M, Dosik H, Groffen J: Evidence of a new chimeric bcr/c-abl mRNA in patients with chronic myelocytic leukemia and the Philadelphia chromosome. N Engl J Med 313:1429-1433, 1985
Wiedmann LM, Karhi KK, Shivji MKK, Rayter SI, Pegram SM, Dowden G, Devan D, Will A, Galton DAG, Chan LC: The correlation of bcr rearrangement of p120 ph1/c-abl expression with morphological analysis of Ph negative CML and other myeloproliferative disease. Blood 71:349-355, 1988
de Klein A, van Kessel AG, Grosveld G, Bartram CR, Hagmeijer A, Bootsma D, Spurr NK, Heisterkamp N, Groffen J, Stephenson JR: A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukemia. Nature 300:765-767, 1982
Knopka JB, Watanabe SM, Witte ON: An alteration of the human c-abl protein in K562 leukemic cells unmasks associated tyrosine kinase activity. Cell 37:1035-1042, 1984
Dhut S, Dorey EL, Horton MA, Ganesan TS, Young BD: Identification of two normal bcr gene products in the cytoplasm. Oncogene 3:561-566, 1988
Harlow E, Lane D: Antibody. A Laboratory Manual. Cold Spring Harbor, New York, Cold Spring Harbor Laboratory Press, 1988
Southern EM: Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503-517, 1975
Chomczynski P, Sacchi N: Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156, 1987
Maller JL: Xenopus oocytes and the biochemistry of cell division. Biochemistry 29:3157-3166, 1990
Lewin B: Driving the cell cycle; M phase kinase, its partners, and substrates. Cell 61:743-752, 1990
Ron D, Zannini M, Levis M, Wickner RB, Hunt LT, Graziani G, Tronick SR, Aaronson SA, Eva A: A region of proto-dbl essential for its transforming activity shows sequence similarity to a yeast cell cycle gene, CDC24, and the human breakpoint cluster gene, bcr. New Biol 3:372-379, 1991
Hartwell LH, Mortimer RK, Culotti J, Culotti M: Genetic control of the cell division cycle in yeast. V. Genetic analysis of cdc mutants. Genetics 74:267-286, 1973
Ohya Y, Miyamoto S, Ohsumi Y, Anraku Y: Calcium sensitive cls4 mutant of Saccharomyces cerevisiae with a defect in bud formation. J Bacteriol 165:28-33, 1986
Bernards A, Rubin CM, Westbrook CA, Paskind M, Baltimore D: The first intron in human c-abl gene is at least 200 kilobases long and is a target for translocations in chronic myelogenous leukemia. Mol Cell Biol 7:3231-3236, 1987
Shtivelman E, Lifshitz B, Gale RP, Roe BA, Canaani E: Alternative splicing of RNAs transcribed from the human abl gene and from the bcr-abl fused gene. Cell 47:277-283, 1986
Rights and permissions
About this article
Cite this article
Miyazaki, Y., Mitsuma, T., Ichida, T. et al. Amplification of BCR Protein Associated with Oncogenesis in Human Hepatocellular Carcinoma. Dig Dis Sci 42, 927–937 (1997). https://doi.org/10.1023/A:1018864414582
Issue Date:
DOI: https://doi.org/10.1023/A:1018864414582