Abstract
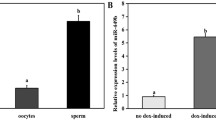
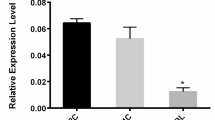
Changes in microRNA (miRNA) levels are present in numerous diseases. Although these changes are particularly noted in male infertility, little is known about the effects of increased miR-16-1 in sperm from infertile men. In this study, we assessed the effects of increased mir-16-1 expression on the developmental process, epigenetic changes, and target gene expressions. IVF embryos, 6 h after insemination, were divided into three groups: control, control negative (CN), and miR-16-1 harboring plasmid microinjection. The developmental rates of these embryos were recorded after 24, 48, 72, and 96 h of culture. The levels of histone H3 lysine 4 tri-methylation (H3K4me3) and histone H3 lysine 27 tri-methylation (H3K27me3) were assessed in the 2-cell and blastocyst stages by immunofluorescence staining. Expression profiles of the miR16-1, Bax, Bcl-2, Suz12, and Kmt2a genes were measured by quantitative real-time polymerase chain reaction (qRT-PCR). There was a significant decrease from the 8-cell stage to the blastocyst stage of embryo development in the miR-16-1 harboring plasmid microinjection group. We observed substantial reductions in the amounts of H3K4me3 and H3K27me3 in the 2-cell and the blastocyst stages in the miR-16-1 harboring plasmid microinjection group (P ≤ 0.05). The miR-16-1 level in the miRNA group was higher than the control group in the 2-cell and the blastocyst stages. There was a significant increase (P ≤ 0.05) in Bax and decreases in Bcl2, Suz12, and Kmt2a following the injection of the miR-16-1 harboring plasmid. These results suggest that a change in miR-16-1 expression can significantly affect embryo development, epigenetic changes, and target gene expressions.







Similar content being viewed by others
References
Wang J, Sauer MV. In vitro fertilization (IVF): a review of 3 decades of clinical innovation and technological advancement. Ther Clin Risk Manag. 2006;2(4):355 364.
Hardy K, Spanos S, Becker D, Iannelli P, Winston R, Stark J. From cell death to embryo arrest: mathematical models of human preimplantation embryo development. Proc Natl Acad Sci. 2001;98(4):1655–60.
Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306(5701):1574–7.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97.
Pasquinelli AE, Hunter S, Bracht J. MicroRNAs: a developing story. Curr Opin Genet Dev. 2005;15(2):200–5.
Aqeilan R, Calin GA, Croce C. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17(2):215–20.
Salas-Huetos A, Blanco J, Vidal F, Godo A, Grossmann M, Pons MC, et al. Spermatozoa from patients with seminal alterations exhibit a differential micro-ribonucleic acid profile. Fertil Steril. 2015;104(3):591–601.
Abu-Halima M, Hammadeh M, Schmitt J, Leidinger P, Keller A, Meese E, et al. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil Steril. 2013;99(5):1249–55. e16.
Kotaja N. MicroRNAs and spermatogenesis. Fertil Steril. 2014;101(6):1552–62.
Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci. 2005;102(39):13944–9.
Haouzi D, Hamamah S. Pertinence of apoptosis markers for the improvement of in vitro fertilization (IVF). Curr Med Chem. 2009;16(15):1905–16.
Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J. 2011;278(10):1598–609.
Ringrose L, Paro R. Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Annu Rev Genet. 2004;38:413–43.
Ringrose L, Ehret H, Paro R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell. 2004;16(4):641–53.
Santos-Rosa H, Schneider R, Bernstein BE, Karabetsou N, Morillon A, Weise C, et al. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol Cell. 2003;12(5):1325–32.
Li B, Howe L, Anderson S, Yates JR, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278(11):8897–903.
Blackledge NP, Klose R. CpG island chromatin: a platform for gene regulation. Epigenetics. 2011;6(2):147–52.
Wong C-M, Wong CC-L, Ng Y-L, Au SL-K, Ko FC-F, Ng IO-L (2011) Transcriptional repressive H3K9 and H3K27 methylations contribute to DNMT1-mediated DNA methylation recovery. PLoS One. 6(2).
Cheng X, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49(14):2999–3008.
Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–26.
Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8(5):532–8.
Liu X, Wang C, Liu W, Li J, Li C, Kou X, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537(7621):558–62.
Li F, Wan M, Zhang B, Peng Y, Zhou Y, Pi C, et al. Bivalent histone modifications and development. Curr Stem Cell Res Ther. 2018;13(2):83–90.
Pasini D, Bracken AP, Jensen MR, Denchi EL, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23(20):4061–71.
Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, et al. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci. 2005;102(24):8603–8.
Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta. 2014;1839(12):1362–72.
Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25(3):338–42.
Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31(9):2305–12.
Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–4.
Brinkman AB, Gu H, Bartels SJ, Zhang Y, Matarese F, Simmer F, et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012;22(6):1128–38.
Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–70.
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–53.
Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39(4):457–66.
Hu J-L, Zhou BO, Zhang R-R, Zhang K-L, Zhou J-Q, Xu G-L. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc Natl Acad Sci. 2009;106(52):22187–92.
Moreno-Moya JM, Vilella F, Simón C. MicroRNA: key gene expression regulators. Fertil Steril. 2014;101(6):1516–23.
Vahdat-Lasemi M, Hosseini S, Jajarmi V, Kazemi B, Salehi M. Intraovarian injection of miR-224 as a marker of polycystic ovarian syndrome declines oocyte competency and embryo development. J Cell Physiol. 2019;234(8):13858–66.
Hosseini S, Dehghani-Mohammadabadi M, Ghafarri Novin M, Haji Molla Hoseini M, Arefian E, Mohammadi Yeganeh S, et al. Toll-like receptor4 as a modulator of fertilization and subsequent pre-implantation development following in vitro maturation in mice. Am J Reprod Immunol. 2017;78(5):e12720.
Tang F, Kaneda M, O’Carroll D, Hajkova P, Barton SC, Sun YA, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21(6):644–8.
Yang Q, Lin J, Liu M, Li R, Tian B, Zhang X, et al. Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Sci Adv. 2016;2(6):e1501482.
Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, et al. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol. 2010;20(3):271–7.
Reza AMMT, Choi YJ, Han SG, Song H, Park C, Hong K, et al. Roles of microRNAs in mammalian reproduction: from the commitment of germ cells to peri-implantation embryos. Biol Rev. 2019;94(2):415–38.
Nikolov S, Lai X, Vera J (2013) MicroRNA regulation, time delay. Enc Syst Biol
Alikani M, Cohen J, Tomkin G, Garrisi GJ, Mack C, Scott RT. Human embryo fragmentation in vitro and its implications for pregnancy and implantation. Fertil Steril. 1999;71(5):836–42.
Pereda J, Cheviakoff S, Croxatto H. Ultrastructure of a 4-cell human embryo developed in vivo. Hum Reprod. 1989;4(6):680–8.
Exley GE, Tang C, McElhinny AS, Warner CM. Expression of caspase and BCL-2 apoptotic family members in mouse preimplantation embryos. Biol Reprod. 1999;61(1):231–9.
Metcalfe AD, Hunter HR, Bloor DJ, Lieberman BA, Picton HM, Leese HJ, et al. Expression of 11 members of the BCL-2 family of apoptosis regulatory molecules during human preimplantation embryo development and fragmentation. Mol Reprod Dev. 2004;68(1):35–50.
Zhang G, Esteve P-O, Chin HG, Terragni J, Dai N, Corrêa IR Jr, et al. Small RNA-mediated DNA (cytosine-5) methyltransferase 1 inhibition leads to aberrant DNA methylation. Nucleic Acids Res. 2015;43(12):6112–24.
Cai B, Ma M, Chen B, Li Z, Abdalla BA, Nie Q, et al. MiR-16-5p targets SESN1 to regulate the p53 signaling pathway, affecting myoblast proliferation and apoptosis, and is involved in myoblast differentiation. Cell Death Dis. 2018;9(3):1–15.
Taheri H, Hosseini S, Salehi M. The relationship between sperm DNA fragmentation and differential expression of human sperm pro-apoptotic miR-15a/16 and anti-apoptotic BCL-2 gene. Iran J Obstet Gynecol Infertility. 2019;22(10):42–8.
Jurisicova A, Latham KE, Casper RF, Varmuza SL. Expression and regulation of genes associated with cell death during murine preimplantation embryo development. Mol Reprod Dev. 1998;51(3):243–53.
Hardy K. Apoptosis in the human embryo. Rev Reprod. 1999;4(3):125–34.
Haghpanah T, Salehi M, Ghaffari Novin M, Masteri Farahani R, Fadaei-Fathabadi F, Dehghani-Mohammadabadi M, et al. Does sperm DNA fragmentation affect the developmental potential and the incidence of apoptosis following blastomere biopsy? Syst Biol Reprod Med. 2016;62(1):1–10.
Wang H, Paulson EE, Ma L, Ross PJ, Schultz RM. Paternal genome rescues mouse preimplantation embryo development in the absence of maternally-recruited EZH2 activity. Epigenetics. 2019;14(1):94–108.
Ross PJ, Ragina NP, Rodriguez RM, Iager AE, Siripattarapravat K, Lopez-Corrales N, et al. Polycomb gene expression and histone H3 lysine 27 trimethylation changes during bovine preimplantation development. Reproduction. 2008;136(6):777–85.
Bogliotti YS, Ross PJ. Mechanisms of histone H3 lysine 27 trimethylation remodeling during early mammalian development. Epigenetics. 2012;7(9):976–81.
Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15(1):57–67.
Gao Y, Hyttel P, Hall VJ. Regulation of H3K27me3 and H3K4me3 during early porcine embryonic development. Mol Reprod Dev. 2010;77(6):540–9.
Polytarchou C, Iliopoulos D, Struhl K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc Natl Acad Sci. 2012;109(36):14470–5.
Chen C-Z, Lodish HF, editors (2005) MicroRNAs as regulators of mammalian hematopoiesis. Semin Immunol; Elsevier.
Shao G-B, Chen J-C, Zhang L-P, Huang P, Lu H-Y, Jin J, et al. Dynamic patterns of histone H3 lysine 4 methyltransferases and demethylases during mouse preimplantation development. In Vitro Cell Dev Biol Anim. 2014;50(7):603–13.
Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95.
Varier RA, Timmers HM. Histone lysine methylation and demethylation pathways in cancer. Biochimica et Biophysica Acta (BBA)-Reviews on. Cancer. 2011;1815(1):75–89.
Xu Q, Xie W. Epigenome in early mammalian development: inheritance, reprogramming and establishment. Trends Cell Biol. 2018;28(3):237–53.
Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537(7621):553–7.
Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537(7621):548–52.
Wu F, Liu Y, Shang M, Yang X, Ding B, Gao J, et al. Differences in H3K4 trimethylation in in vivo and in vitro fertilization mouse preimplantation embryos. Genet Mol Res. 2012;11(2):1099–108.
Zhang M, Wang F, Kou Z, Zhang Y, Gao S. Defective chromatin structure in somatic cell cloned mouse embryos. J Biol Chem. 2009;284(37):24981–7.
Kim H-G, Kim DJ, Li S, Lee KY, Li X, Bode AM, et al. Polycomb (PcG) proteins, BMI1 and SUZ12, regulate arsenic-induced cell transformation. J Biol Chem. 2012;287(38):31920–8.
Faust C, Lawson KA, Schork NJ, Thiel B, Magnuson T. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development. 1998;125(22):4495–506.
Zheng H, Huang B, Zhang B, Xiang Y, Du Z, Xu Q, et al. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol Cell. 2016;63(6):1066–79.
Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol. 2008;28(5):1862–72.
Wu X, Gong Y, Yue J, Qiang B, Yuan J, Peng X. Cooperation between EZH2, NSPc1-mediated histone H2A ubiquitination and Dnmt1 in HOX gene silencing. Nucleic Acids Res. 2008;36(11):3590–9.
Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–4.
Benjamin DY, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378(6556):505–8.
Fischer CP, Kayisili U, Taylor HS. HOXA10 expression is decreased in endometrium of women with adenomyosis. Fertil Steril. 2011;95(3):1133–6.
Sakkas D, Lu C, Zulfikaroglu E, Neuber E, Taylor HS. A soluble molecule secreted by human blastocysts modulates regulation of HOXA10 expression in an epithelial endometrial cell line. Fertil Steril. 2003;80(5):1169–74.
Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;11(1):5.
Yan J, Guo X, Xia J, Shan T, Gu C, Liang Z, et al. MiR-148a regulates MEG3 in gastric cancer by targeting DNA methyltransferase 1. Med Oncol. 2014;31(3):879.
Jermann P, Hoerner L, Burger L, Schübeler D. Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc Natl Acad Sci. 2014;111(33):E3415–E21.
Lynch MD, Smith AJ, De Gobbi M, Flenley M, Hughes JR, Vernimmen D, et al. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2012;31(2):317–29.
Reddington JP, Perricone SM, Nestor CE, Reichmann J, Youngson NA, Suzuki M, et al. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol. 2013;14(3):R25.
Acknowledgements
We deeply appreciate all those who assisted with the completion of this article. We would like to thank Ms. Dehghan for her assistance with the genetic analyses and Dr. Bakhtari for his comments that greatly improved the immunofluorescence staining.
Funding
This research was supported by the Cellular and Molecular Research Center of Shahid Beheshti University of Medical Sciences, and part of the cost of this research was provided by the School of Veterinary Medicine of Shiraz University. Also, part of this research was reinforced by the Department of Genetics of Shahid Beheshti University of Medical Sciences.
Author information
Authors and Affiliations
Contributions
Experimental design and data analysis were carried out by M. Salehi and the practical steps in the laboratory were carried out by M. Kiani and S. Shahidi. Samira Mohammadi Yeganeh was involved in the genetics department and A. Mogheiseh contributed in revisions and modifications for the final manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kiani, M., Salehi, M., Mogheiseh, A. et al. The Effect of Increased miR-16-1 Levels in Mouse Embryos on Epigenetic Modification, Target Gene Expression, and Developmental Processes. Reprod. Sci. 27, 2197–2210 (2020). https://doi.org/10.1007/s43032-020-00240-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-020-00240-4