Abstract
The problem of identification of single crop fields is a challenge when single date optical remote sensing image is used. The use of temporal images solves this problem. However, issues like cloud cover in optical images influence accuracy of results. Microwave data, which penetrate through the atmosphere, solve this problem. The existence of mixed pixels in satellite images and nonlinearity in image classification is also overlooked. These issues were considered and worked on by integrating C band RISAT-1 with Formosat-2 temporal images and using possibilistic c-means classifier with similarity and dissimilarity norms to identify late transplanted paddy (Oryza sativa) fields in Haridwar District of India. Three datasets in different temporal combinations of microwave and optical images were classified for various similarity and dissimilarity norms for different values of weighted constant. Favorable results were achieved for Manhattan and mean absolute difference norm at weighted constant m = 1.3. Classification of late transplanted paddy for datasets containing multiple RISAT-1 and single Formosat-2 images with transplanting, growth stages was found to yield best results as compared to other combination of temporal images.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Identification of a single crop has an advantage for the government as they can undertake different policies for the masses as well as regulate import and export strategy. Crop type maps often come to rescue of national and regional agricultural. They provide information to facilitate water resource planning for irrigation [1, 2], crop yield assessment and forecasting [3, 4] as well as mapping soil productivity [5]. A lot of fluctuations have been seen in the grains market especially in the last decade. The production of wheat and rice dropped from 63 to 16 MT between 2002 and 2007 [6]. This situation was of major concern for the national food security. Hence, for efficient analysis of such conditions, monitoring single crop for its yield, acreage and agricultural pattern is essential [7].
Orthodox methods to compose crop type maps are based on ground surveying and census and record keeping [8]. These methods lack standardization. In order to standardize, the continuous nature of collecting information using remotely sensing satellite has proven efficient [9,10,11]. Satellite images obtained from different sensors and of different time durations can be clubbed together to obtain datasets with relatively low spectral dimensions. Many studies have exploited the use of optical images for carrying out crop-based analysis [12,13,14,15].
One major concern related to the optical data still remains the validity of how accurately the atmospheric corrections are done. This issue is confidently handled by microwave remote sensing [16,17,18]. Crop identification can be successfully carried out by classifying temporal satellite images as reflectance corresponds to crop phenology and single date image analysis is a challenge [19]. Pixel-based hard classification technique where spectral mixing at class boundaries does not exist has been traditionally carried out in crop studies [13, 20]. In such studies, nonlinearity of classes goes unnoticed. The present study tries to overcome this problem of hard classification by using soft classification technique for handling mixed pixels using temporal normalized difference vegetation index (NDVI) obtained from Formosat-2 and temporal synthetic aperture radar (SAR) images obtained from Radar Imaging Satellite-1 (RISAT-1) for classification paddy (Oryza sativa).
The complete growth cycle of paddy is divided into five stages and is observed in the time frame starting from end of June to the start of November. These stages include the transplanting period; seedling development stage; ear differentiation period; the heading period; and the maturation period where the rice plant is matured and ready for harvesting [21]. It is observed that long growing season is favorable for high paddy yields, and hence, the transplanting season starts at an early date as compared to the previous growth cycles. The life cycle of paddy is around 120–150 days, but a variation is observed as the paddy type changes. The spectral signatures at crop growth stages in the time domain can help in discriminating various crops and vegetation patterns [22]. This phenological aspect was used to map paddy fields through multi-temporal datasets.
The objective was to classify paddy fields over a region and estimate the regions with maximum possibility of paddy cultivation. The supervised classification was chosen over unsupervised one as it provided the user to manipulate the pixel’s spectral values and not rely on the clustering patterns generated by algorithms. The spatial resolution of satellite image is directly proportional to the values captured in the pixels. Coarser the spatial resolution higher is the chance that the pixel is a part of more land cover types; these pixels are termed as mixed pixels. In traditional hard classification techniques, algorithms assign pixels to specific land cover types which results in loss of information. To handle the mixed pixel issue in the classification, possibilistic c-means (PCM) classifier was selected over fuzzy c-means (FCM) classifier as membership values of PCM are a measure of ‘degree of belonging’ [23], while that of FCM is ‘degree of sharing’ [24]. In PCM, the clustering problem is drafted in the possibility domain where the resultant partitions are interpreted as possibilistic partition.
The major objective of the research was to soft classify the bi-sensor temporal images and compare between the norms for the best separation of linear classes. Other objectives include: (1) to evaluate combined bi-sensor datasets for better extraction of paddy fields, (2) to identify best date combination of bi-sensor datasets for identification of paddy fields and (3) to compare similarity and dissimilarity norms via PCM classifier.
2 Indices and measures
2.1 Normalized difference vegetation index (NDVI)
In order to reduce the spectral dimensionality of the dataset, vegetation indices were used. The NDVI band ratio was proposed by Kriegler et al. [25]. It was observed that the vegetation pigments have high absorptivity in the red spectral wavelength and high reflectance in the near infrared wavelength. This observation was successfully drafted into a band ratio where Red and NIR bands were used to reduce dimension of dataset. The NDVI is calculated using Eq. (1):
where \(\rho_{{{\text{NIR}}}}\) represents reflectance at near infrared band and \(\rho_{{{\text{RED}}}}\) represents reflectance at red band.
2.2 Possibilistic c-means (PCM) classification method
PCM classifier generates membership values which are interpreted as degree of belongingness or typicality [26]. The actual feature classes should have high membership values as compared to the values associated with unrepresentative features. The objective function is expressed in Eq. (2):
where C is the number of classes, N is the number of pixels, m is the weighted constant, and \(\eta_{i}\) (scale or resolution parameter) are suitable positive numbers. The first term demands that the distance from the feature vector to the prototypes be as low as possible, whereas the second term forces \(u_{ij}\) (fuzzy membership value) to be as large as possible to avoid the trivial solution. The resolution parameter is calculated as in Eq. (3):
The fuzzy membership value \(u_{ij}\) is calculated from Eq. (4):
2.3 Similarity and dissimilarity measures
2.3.1 Similarity measures
Similarity measure between two sequences is a measure that quantifies dependencies between them. The similarity measures of cosine and correlation are applied with PCM classifier.
Cosine This similarity norm measures the cosine angle between two vectors of an inner product space. It is mentioned in Eq. (5):
Correlation Pearson’s correlation coefficient (r) is used to measure the similarity between the two items. The same is formulated in Eq. (6):
2.3.2 Dissimilarity measures
This measure between two sequences quantifies the independency between them. The dissimilarity measure D is considered a metric if it produces a higher value as corresponding values in the sequence become less independent. A total of ten dissimilarity norms of Bray Curtis, chessboard, Manhattan, Canberra, Euclidean, mean absolute distance, median-absolute distance and normalized squared Euclidean are evaluated in the PCM classifier [27].
Bray Curtis This norm is a statistic used to quantify the compositional dissimilarity between two sites based on counts on each site [28]. Equation (7) describes this norm.
Chessboard Also known as the Chebyshev distance, this metric distance is defined on a vector space where the distance between the two vectors is the greatest of their distances along any coordinate dimension. Equation (8) gives the chessboard norm.
Manhattan This norm is the sum of absolute intensity differences and is one of the oldest norms. The generalized equation used is shown in Eq. (9).
Canberra This norm is the weighted version of Manhattan distance. It is a numerical measure of point pairs in vector space. The formula is stated in Eq. (10).
Euclidean Euclidean distance between two points is calculated as square root of the sum of the squares of the difference between the corresponding points. Equation (11) explains the Euclidean distance. The Mahalanobis and diagonal Mahalanobis norms are calculated as per Eq. (12).
where \(A^{ - 1}\) is both variance–covariance and diagonal variance–covariance.
Mean Absolute Difference This norm measures the mean of the absolute deviation from the central point. It is the summary statistics of variability. Equation (13) discussed this norm.
Median-Absolute Difference This dissimilarity norm reduces the effect of impulse noise on the calculated images. The formula for this norm is represented in Eq. (14).
Normalized Squared Euclidean This norm normalizes the measure with respect to the image contrast. In the calculation of correlation coefficient, scale normalization is performed once after calculating the inner product of the normalized intensities. Equation (15) describes this norm.
2.4 Backscattering coefficient
The backscatter coefficient (\({\sigma }^{0}\)) is defined as the differential scattering cross section per unit volume for a scattering angle of 180′. Measurements of this quantity involve the projection of a pulsed ultrasound beam into a volume containing the medium of interest and monitoring echo signals due to scattering. The formula used for calculating backscattering coefficient for RISAT-1 data is given in Eq. (16) [29].
where \({\text{DN}}_{{\text{p}}}\) is the digital number for the pixel p, \(K_{{{\text{dB}}}}\) is the calibration constant in dB, \(i_{p} \; {\text{and }}\;i_{{{\text{center}}}}\) are the incidence angle for pixel p and center of the scene, respectively. The value of \({\text{DN}}_{{\text{p}}}\) is calculated using Eq. (17).
where \({\text{DNI}}_{{\text{p}}}\) is DN value of in-phase (real channel) component and \({\text{DNQ}}_{{\text{p}}}\) is DN value of quadrature (imaginary channel) component.
3 Study area and data used
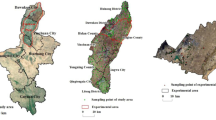
The study area is situated on the east side of Haridwar, Uttarakhand, India, toward national highway 74 as seen in Fig. 1. The central latitude and longitude of the area are 29°52′20.3124″N and 78°10′25.0998″E. River Ganges flows through the district, and hence, the land here is fertile and conducive for agriculture. The major crops cultivated in this region include wheat, rice, sugarcane, mustard, groundnuts and fruits like mangoes and litchis. The temperature in summer ranges from 25 to 44 °C, while that in winter ranges from − 2 °C to 24 °C. The area of the city is 12.3 km2 (Table 1).
In most of the researches carried out, optical data were used extensively for crop mapping. In India, the monsoon season coincides with the transplanting season for paddy crop. Due to this, the remotely sensed data have cloud cover which makes it difficult to understand and exploit the pixel values. The occurrence of atmospheric disturbances, cloud cover, creates gaps in temporal data and decreases the accuracy of results [30]. Data used for this research work include four satellite images obtained from RISAT-1 (two images) and Formosat-2 (two images). RISAT-1 images are microwave images in the C band (5.35 GHz) frequency range with dual polarization HH and HV in Medium Resolution ScanSAR (MRS) mode. The electromagnetic radiation is a combination of electric and magnetic waves in which the electric field dictates the direction of propagation. When the receiving antenna points in the same direction of that of propagation, best results are achieved. Hence, if the propagation of waves is in the horizontal domain and the antenna also points in the same direction, the polarization achieved is termed as HH.
The study uses the HH polarization as it was found suitable for paddy monitoring [21, 31]. Based on their studies in the Zhaoqing test site, they established an experimental backscattering model in which the backscattering was shown as a function of time using cubic polynomial. This study was taken into account while short listing the polarization to be used. The spatial resolution in MRS mode for RISAT-1 is 18 m, while that of Formosat-2 is 8 m. As crop mapping cannot be determined accurately using single date image [19], temporal data were used. The temporal dates of RISAT-1 data were 27 June 2014 and 09 July 2014, while that of Formosat-2 were 10 August 2014 and 25 September 2014 as seen in Fig. 2. Ground truth was collected with the help of global positioning system (GPS) points. The survey dates were 19 and 20 October 2014. These data were used for both the training of supervised classification classifier as well as for testing. Of the total points collected, 80% of the points were used for training, while 20% were used for testing purposes. Supervised soft classification was carried on with the JAVA-based sub-pixel multi-spectral image classifier (SMIC) tool with the PCM classifier and the similarity and dissimilarity distance norms. Three temporal datasets were assembled using the four date images in combination of the microwave and optical bands. Dataset one contained two microwave bands dated 27 June and 09 July and one optical band dated 10 August. Dataset two had one microwave band dated 07 July and two optical bands dated 10 August and 25 September, while dataset three had all the four bands (refer Table 2). It was important to select the best date combination to achieve high accuracy. Three-day combination gave better results as compared to single date image or two-date combination [32]. These datasets were constructed after the backscattering coefficients of the microwave data were calculated. NDVI index was calculated using optical images of Formosat-2.
4 Methodology and approach
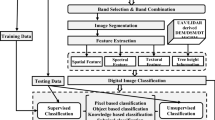
Bi-sensor approach was used to carry out the research. This was done to understand if data fusion for classification would yield desired results. Methodology followed is presented in Fig. 3.
RISAT-1 images dated 27 June and 9 July 2014 were geometrically corrected to be in accordance with Formosat-2 temporal images dated 10 August and 25 September 2014. RISAT-1 images were processed to obtain backscattering images using formula 16 mentioned above and resampled to 8 m to match the spatial resolution of Formosat-2. NDVI image was obtained from Formosat-2 temporal images. The backscattered images and NDVI images were linearly stretched to obtain 8 bit images with pixel values ranging from 0 to 255. This was done as the SMIC (sub-pixel land cover mapping image classifier), a JAVA-based image processing package [33] which supports 8 bit imagery was used for classification. The linear stretching would overcome the difference created by decibel values of backscattering images and NDVI values bringing uniformity in analysis.
4.1 Dataset generation
Three datasets were generated using temporal images. These datasets were obtained by stacking backscatter images and NDVI images in order of the date of acquisition of images. The stacking of these bands and the dates is as seen in Table 2.
The ground data which were utilized for the training and testing of sites were collected on 19 and 20 October 2014 through field survey. These data were used to accurately locate the pure pixels, i.e., to identify the paddy fields in the region of study. Pure pixels are needed for training as fields in India are closely situated and spectral mixing of signature may result in nonlinearity in classes. The growth cycle under study belongs to the kharif season which is monsoon dependent in India. Typically, it takes anywhere between 120 and 150 days for paddy to reach maturity. The collection of ground data was close to the late transplanted paddy, and hence, identification of this single class was carried out. The target class of late transplanted paddy was to be isolated from nearby paddy fields which were basically early transplanted paddy and nearing maturity.
Nearly 15 pure pixels from six different paddy field sites were used to carry out supervised kernel-based PCM using SMIC package. Different types of similarity and dissimilarity based distance norm kernels were used with various values of weighted constant ‘m’ ranging from 1.5 to 3 for each kernel.
5 Results and discussion
Agricultural fields change temporally hence to classify these changes temporal images are used. As quantitative evaluation of the fields was not possible, the evaluation was done based on the membership values of PCM for varying values of ‘m.’ The fundamental of soft classification which says inter-class membership variance should be more and intra-class membership variance should be low [34] was used considering factors like optimized weighted constant ‘m’, temporal combination of images and similarity and dissimilarity-based distance norms.
Figures 4, 5 and 6 show the classified outcomes of dataset 1, 2 and 3 for late transplanted paddy for optimized weighted constants as seen in Tables 3, 4 and 5, respectively.
Late transplanted paddy extracted for dataset 1 using a Bray Curtis, b Canberra, c chessboard, d correlation, e cosine, f diagonal variance–covariance, g Euclidean, h mean absolute difference, i Manhattan, j median absolute difference, k normalized square Euclidean, l variance–covariance for optimized weighted constants
Late transplanted paddy extracted for dataset 2 using a Bray Curtis, b Canberra, c chessboard, d correlation, e cosine, f diagonal variance–covariance, g Euclidean, h mean absolute difference, i Manhattan, j median absolute difference, k normalized square Euclidean, l variance–covariance for optimized weighted constants
Late transplanted paddy extracted for dataset 3 using a Bray Curtis, b Canberra, c chessboard, d correlation, e cosine, f diagonal variance–covariance, g Euclidean, h mean absolute difference, i Manhattan, j median absolute difference, k normalized square Euclidean, l variance–covariance for optimized weighted constants
The membership values for different testing sites were noted for target class (late transplanted paddy) and non-target classes (early transplanted paddy and shallow water) for all the three datasets. The target and non-target classes selected were based on the temporal images used. Paddy fields are filled with water during the transplantation stage. This often leads to misclassification when class of shallow water is present in the study area resulting in similar backscattering values. The maximum membership variance for corresponding weighted constant will result in optimized weighted constant, whereas the maximum variance in membership for optimized weighted constants for different datasets will result in best temporal combination of dataset as nonlinearity was handled best for that combination of distance norm, temporal combination and weighted constant.
Tables 3, 4 and 5 clearly depict the membership values for unbiased testing sites for dataset 1, 2 and 3, respectively. The optimized weighted constant for which the difference of membership values between the target and non-target sites is mentioned in the tables. For dataset 1 with temporal images of June, July and August show that for optimized ‘m’ = 1.3, mean absolute difference and Manhattan distance norms yielded highest mean membership values for late transplanted paddy class of 253 of all unbiased testing sites. These norms provided the maximum mean membership values for target class (refer Table 3). For dataset 2 (refer Table 4) with temporal images of July, August and September maximum mean membership value of target class was observed for diagonal variance–covariance norm at 254 with optimized ‘m’ = 2.3.
Whereas (refer Table 5) for dataset 3 which consists of all temporal images of June, July, August and September maximum mean membership value for target class calculated from unbiased sites was observed for variance–covariance norm at 253 where optimized ‘m’ was 2.1
Graphs shown in Fig. 7 are plotted for the difference between the membership values of late transplanted paddy and early transplanted paddy for datasets 1, 2 and 3, respectively. Figure 8 represents the graphs for difference of membership values between late transplanted paddy and shallow water for 12 norms at various values of weighted constant. These differences are based on the 8 bit classified images obtained using SMIC classifier. The average difference of unbiased sites is plotted against weighted constants ‘m.’
Table 6 gives an overview of the best combination of dataset, distance norm and corresponding optimized weighted constant for which the classification of late transplanted paddy yielded best statistical results.
6 Conclusions
A total of 12 similarity and dissimilarity norms were tested over 3 datasets with temporal resolution of 3 and 4 dates for better extraction of single class that is late transplanted paddy fields. It was observed that the weighted constant ‘m’ played a significant role in suppressing non-target classes when single class classification was carried out. The suppression of non-target classes was not uniform as difference between the membership values of the target and non-target classes were uniform as microwave and optical images characteristics came into picture. In some cases, the difference between the membership values of late transplanted paddy and early transplanted paddy was maximum, while in few cases, the difference between late transplanted paddy and shallow water was maximum.
It was observed that the PCM classifier could identify and extract single class and suppress other classes and also solve the problem of mixed pixels to a larger extent when datasets were compared. Statistically, it was found that the dataset containing two dates of microwave data and one date of optical data produced best results for the norms mean absolute difference and Manhattan at weighted constant value ‘m = 1.3.’ However, visual interpretation suggested that the dataset containing two dates of microwave data and two dates of optical data had better results for the norm variance–covariance at ‘m = 2.1’ as seen in Figs. 4, 5 and 6.
It was thus concluded that the PCM classifier produces satisfactory results for single class extraction and also manages mixed pixels to suppress non-target classes for dataset containing more microwave temporal images than optical images. The noise observed in the classification images can be handled by applying speckle filter before the classification process. From the results obtained, it can be concluded that the use of multi-date microwave data when integrated with optical data produces better results as compared to the use of single date microwave data with optical data in temporal scenarios for crop identification and mapping.
References
Abou-EL-Magd I, Tanton TW (2003) Improvements in land use mapping for irrigated agriculture from satellite sensor data using a multi-stage maximum likelihood classification. Int J Remote Sens 24(21):4197–4206
Zhong L, Hawkins T, Biging G, Gong P (2011) A phenology-based approach to map crop types in the San Joaquin Valley, California. Int J Remote Sens 32(22):7777–7804
Doraiswamy PC, Moulin S, Cook PW, Stern A (2003) Crop yield assessment from remote sensing. Photogramm Eng Remote Sensing 69(6):665–674
Bolton DK, Friedl MA (2013) Forecasting crop yield using remotely sensed vegetation indices and crop phenology metrics. Agric For Meteorol 173:74–84
Anderson-Cook CM, Alley MM, Roygard JKF, Khosla R, Noble RB, Doolittle JA (2002) Differentiating soil types using electromagnetic conductivity and crop yield maps. Soil Sci Soc Am J 66(5):1562–1570
Nandakumar T, Ganguly K, Sharma P, Gulati A (2010) Food and nutrition security status in India: Opportunities for investment partnerships.
Panigrahy RK, Ray SS, Panigrahy S (2009) Study on the utility of IRS-P6 AWIFS SWIR band for crop discrimination and classification. J Indian Soc Remote Sensing 37(2):325–333
Nandan R, Kamboj A, Kumar A, Kumar AS, Reddy KV (2016) Formosat-2 with Landsat-8 temporal-multispectral data for wheat crop identification using Hypertangent Kernel based Possibilistic classifier. J Geomatics 10(1)
Panigrahy S, Sharma SA (1997) Mapping of crop rotation using multidate Indian remote sensing satellite digital data. ISPRS J Photogramm Remote Sensing 52(2):85–91
Salmon JM, Friedl MA, Frolking S, Wisser D, Douglas EM (2015) Global rain-fed, irrigated, and paddy croplands: a new high resolution map derived from remote sensing, crop inventories and climate data. Int J Appl Earth Obs Geoinf 38:321–334
Pacheco A, McNairn H (2010) Evaluating multispectral remote sensing and spectral unmixing analysis for crop residue mapping. Remote Sens Environ 114(10):2219–2228
Boschetti M, Stroppiana D, Brivio PA, Bocchi S (2009) Multi-year monitoring of rice crop phenology through time series analysis of MODIS images. Int J Remote Sens 30(18):4643–4662
Pan Y, Li L, Zhang J, Liang S, Zhu X, Sulla-Menashe D (2012) Winter wheat area estimation from MODIS-EVI time series data using the Crop Proportion Phenology Index. Remote Sens Environ 119:232–242
Sakamoto T, Wardlow BD, Gitelson AA, Verma SB, Suyker AE, Arkebauer TJ (2010) A two-step filtering approach for detecting maize and soybean phenology with time-series MODIS data. Remote Sens Environ 114(10):2146–2159
Wu B, Meng J, Li Q, Yan N, Du X, Zhang M (2014) Remote sensing-based global crop monitoring: experiences with China's CropWatch system. Int J Digits Earth 7(2):113–137
Clevers JGPW, Van Leeuwen HJC (1996) Combined use of optical and microwave remote sensing data for crop growth monitoring. Remote Sens Environ 56(1):42–51
Oza SR, Panigrahy S, Parihar JS (2008) Concurrent use of active and passive microwave remote sensing data for monitoring of rice crop. Int J Appl Earth Obs Geoinf 10(3):296–304
Soria-Ruiz J, Fernandez-Ordonez Y, McNairn H, Pei-Gee PH (2009) Corn monitoring and crop yield using optical and microwave remote sensing. Geosci Remote Sensing 598
Wardlow BD, Egbert SL, Kastens JH (2007) Analysis of time-series MODIS 250 m vegetation index data for crop classification in the US Central Great Plains. Remote Sens Environ 108(3):290–310
Lohmann P, Soergel U, Tavakkoli M, Farghaly D (2009) Multi-temporal classification for crop discrimination using TerraSAR-X spotlight images. Proc IntArchPhRS 38
Shao Y, Fan X, Liu H, Xiao J, Ross S, Brisco B, Staples G (2001) Rice monitoring and production estimation using multitemporal RADARSAT. Remote Sens Environ 76(3):310–325
El Hajj M, Bégué A, Guillaume S (2007) Multi-source information fusion: monitoring sugarcane harvest using multi-temporal images, crop growth modelling, and expert knowledge. In: 2007 international workshop on the analysis of multi-temporal remote sensing images. IEEE, pp 1–6
Chawla S (2010) Possibilistic c-means-spatial contextual information based sub-pixel classification approach for multi-spectral data. University of Twente Faculty of Geo-Information and Earth Observation (ITC)
Dunn JC (1973) A fuzzy relative of the ISODATA process and its use in detecting compact well-separated clusters
Kriegler FJ (1969) Preprocessing transformations and their effects on multispectral recognition. In: Proceedings of the sixth international symposium on remote sensing of the environment, University of Michigan, Ann Arbor, Michigan, pp 97–131
Krishnapuram R, Keller JM (1993) A possibilistic approach to clustering. IEEE Trans Fuzzy Syst 1(2):98–110
Tyagi U, Kumar A (2015) Similarity and dissimilarity measures with fuzzy classifiers: SMIC tool. In: 9th international conference on ASEICT.
Bray JR, Curtis JT (1957) An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27(4):325–349
Rao SS, Sahadevan DK, Wadodkar MR, Nagaraju MSS, Chattaraj S, Joseph W, Rajankar P, Sengupta T, Venugopalan MV, Das SN, Joshi AK (2014) Soil moisture model with multi angle and multi polarisation risat-1 Data. ISPRS Ann Photogramm Remote Sensing Spatial Inf Sci 2(8):145
Steven MD, Malthus TJ, Baret F, Xu H, Chopping MJ (2003) Intercalibration of vegetation indices from different sensor systems. Remote Sens Environ 88(4):412–422
Kumari M (2015) C-Band RISAT-1 data for crop growth assessment of rice. Asian J Geoinform 15(1)
Murthy CS, Raju PV, Badrinath KVS (2003) Classification of wheat crop with multi-temporal images: performance of maximum likelihood and artificial neural networks. Int J Remote Sens 24(23):4871–4890
Kumar A, Ghosh SK, Dadhwal VK (2006) Sub-pixel land cover mapping: SMIC system. In: ISPRS international symposium on geospatial databases for sustainable development, Goa, India
Tripathi RN, Kumar R, Kumar A, Kumar AS (2017) Wheat monitoring by using kernel based possibilistic c-means classifier: Bi-sensor temporal multi-spectral data. J Indian Soc Remote Sensing 45(6):1005–1014
Acknowledgement
The authors would like to thank Sponsors the National Space Organization, National Applied Research Laboratories and jointly supported by the Chinese Taipei Society of Photogrammetry and Remote Sensing and the Center for Space and Remote Sensing Research, National Central University of Taiwan for providing Formosat-2 temporal images of Haridwar District, Uttarakhand, India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Deshpande, M.M., Kumar, A. & Singh, T.P. Integration of C band SAR and optical temporal data for identification of paddy fields. SN Appl. Sci. 2, 990 (2020). https://doi.org/10.1007/s42452-020-2786-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-020-2786-0