Abstract
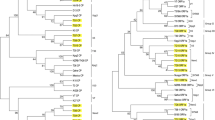
Citrus tristeza virus (CTV) infected plant samples from the North Eastern states of India namely Arunachal Pradesh (AR1), Assam (AS1), Nagaland (NL1), Meghalaya (M1), Sikkim (SK1) and Tripura (TR1) were examined through double antibody sandwich enzyme linked immunosorbent assay and reverse transcriptase polymerase chain reaction using coat protein (CP) gene specific primers, yielding amplicon of 672 base pair for all the isolates. The sequence analysis revealed nucleotide (nt) and amino acid (aa) sequence similarity with other exotic CTV isolates. AR1 and AS1 shared similarity with T3 isolate (Florida) at 98–99% and 95–99% for nt-aa respectively, whereas, M1 showed 99% similarity with stem pitting isolate B165 (Bangalore). SK1 and NL1 were 95–97% and 97–99% similar with Hawaiian isolate HA at nt-aa level. TR1 showed 97% homology with seedling yellowing Israeli isolate VT. The CP gene sequence based phylogenetic analysis grouped present isolates into four clades with reference severe isolates (i.e. VT, B165, HA and T3), indicating their probable evolution. TR1 and NL1 are the putative recombinant isolates and ASI and SK1 were the major and minor parents respectively. This might be due to the conserved nature of CP gene. Moreover, the CP gene sequences of present isolates showing nucleotide identity to the VT, T3 and B165 isolates also suggest the prevalence of severe CTV isolates in Khasi mandarin growing areas comes under North East (NE) region of India.




Similar content being viewed by others
References
Ahlawat YS (2005) Virus and virus—like diseases affecting citrus in India—a step forward to management of die—back complex. Indian Phytopathol 3:257–268
Ahlawat YS (2007) Citrus decline: problems and prevention. Indian Phytopathol 60(1):1–12
Anonymous (2014) Horticultural database. National Horticultural Board, Gurgaon
Bar-Joseph M, Dawson WO (2008) Citrus tristeza virus. Encycl Virol 1:520–525
Barzegar A, Sohi HH, Rahimian H (2006) Characterization of Citrus tristeza virus isolates in northern Iran. J Gen Plant Pathol 72:46–51
Biswas KK (2008) Molecular Diagnosis of Citrus tristeza virus in Mandarin (Citrus reticulata) orchard of Dargiling hills of west Bengal. Indian J Virol 19(1):26–31
Biswas KK (2010) Molecular characterization of Citrus tristeza virus isolates from the North Eastern Himalayan region of India. Arch Virol 155:959–963
Biswas KK, Tarafdar A, Sharma SK, Singh JK, Dwivedi S, Biswas K, Jayakumar BK (2014) Current status of Citrus tristeza virus incidence and its spatial distribution in citrus growing geographical zones of India. Indian J Agric Sci 2:184–189
Boni MF, Posada D, Feldman MW (2007) An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047
Borah M, Nath PD, Saikia AK (2012) Serological detection of Citrus tristeza virus affecting citrus tree species in Assam. Indian Phytopathol 3:289–293
Borah M, Nath PD, Saikia AK (2014) Biological and serological techniques for detection of Citrus tristeza virus affecting Citrus species of Assam, India. Afr J Agric Res 52:3804–3810
Clark MF, Adams AN (1977) Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J Gen Virol 34:475–483
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucl Acids Res 16(22):10881–10890
Dawson TE, Mooney PA (2000) Evidence for trifoliate resistance breaking isolates of Citrus tristeza virus in New Zealand. In: da Graca JV, Lee RF, Yokomi RK (eds) Proceeding of 14th conference international organization citrus virology. International Organization of Citrus Virologists, Riverside, pp 69–76
Food and Agriculture Organization (FAO) Statistical year book (2017) Published by Food and Agriculture Organization of the United Nations. www.fao.org/faostat/en/#data/QC
Ghosh SP, Singh RB (1993) Citrus in south Asia. FAO/RAPA publication no. 1993/24. Bangkok, Thailand, p 70
Gibbs MJ, Armstrong JS, Gibbs AJ (2000) Sister-Scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582
Guan-Wei W, Min T, Guo-Ping W, Feng-Yin J, Zuo-Kun Y, Li-Jing C, Ni H (2015) Genetic diversity and evolution of two capsid protein genes of citrus tristeza virus isolates from China. Arch Virol 160:787–794
Harper SJ (2013) Citrus tristeza virus: evolution of complex and varied genotypic groups. Front Microbiol 4:93. https://doi.org/10.3389/fmicb.2013.00093
Harper SJ, Dawson TE, Pearson MN (2009) Complete genome sequences of two distinct and diverse Citrus tristeza virus isolates from New Zealand. Arch Virol 154(9):1505–1510
Harper SJ, Dawson TE, Pearson MN (2010) Isolates of Citrus tristeza virus that overcome Poncirus trifoliata resistance comprise a novel strain. Arch Virol 155:471–480
Hilf ME, Mavrodieva VA, Garnsey SM (2005) Genetic marker analysis of a global collection of isolates of Citrus tristeza virus: characterization and distribution of CTV genotypes and association with symptoms. Phytopathology 95:909–917
Holmes DS, Quigley M (1981) A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem 114:193–197
Jiang B, Hong N, Wang GP, Hu J, Zhang JK, Wang CX, Liu Y, Fan XD (2008) Characterization of Citrus tristeza virus strains from southern China based on analysis of restriction patterns and sequences of their coat protein genes. Virus Genes 37:185–192
Karasev AV, Boyko VP, Gowda S, Nikolaeva OV, Hilf ME, Koonin EV, Nibblet CL, Cline K, Gumpf DJ, Lee RF, Garnsey SM, Dawson WO (1995) Complete sequence of the Citrus tristeza virus RNA genome. Virology 208:511–520
Kishore K, Rahman H, Kalita H, Pandey B, Minika N (2010) Prevalence of Citrus tristeza virus in mandarin of Sikkim Himalayan Region. Ind J Virol 21:140–143
Lbida B, Bennani A, Serrhini MN, Zemzami M (2005) Biological, serological and molecular characterization of three isolates of Citrus tristeza Closterovirus introduced into Morocco. EPPO Bull 35:511–517
Martin S, Sambade A, Rubio L, Vives MC, Moya P, Guerri J, Elena SF, Moreno P (2009) Contribution of recombination and selection to molecular evolution of Citrus tristeza virus. J Gen Virol 90(1):527–538
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:1–5
Mawassi M, Mietkiewska E, Gofman R, Yang G, Bar-Joseph M (1996) Unusual sequence relationships between two isolates of citrus tristeza virus. J Gen Virol 77:2359–2364
Melzer MJ, Borth WB, Sether DM, Ferreira S, Gonsalves D, Hu JS (2010) Genetic diversity and evidence for recent modular recombination in Hawaiian Citrus tristeza virus. Virus Genes 40:111–118
Moreno P, Ambros S, Albiach-Marti MR, Guerri J, Pena L (2008) Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Mol Plant Pathol 9:251–268
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York, p 126
Niblett CL, Genc H, Cevik B, Halbert S, Brown L, Nolasco G, Bonacalza B, Manjunath KL, Febres VJ, Pappu HR, Lee RF (2000) Progress on strain differentiation of citrus tristeza virus and its application to the epidemiology of citrus tristeza disease. Virus Res 71:97–106
Nolasco G, Santos C, Silva G, Fonseca F (2009) Development of an asymmetric PCR-ELISA typing method for Citrus tristeza virus based on the coat protein gene. J Virol Methods 155:97–108
Pappu HR, Pappu SS, Manjunath KL, Lee RF, Niblett CL (1993a) Molecular characterization of a structural epitope that is largely conserved among severe isolates of a plant virus. Proc Natl Acad Sci USA 90:3641–3644
Pappu HR, Pappu SS, Niblett CL, Lee R, Civerolo E (1993b) Comparative sequence analysis of the coat protein of biologically distinct citrus tristeza Closterovirus isolates. Virus Genes 7:255–264
Rocha-Pena MA, Lee RF, Lastra R, Niblett CL, Ochoa- Corona FM, Garnsey SM, Yokomi RK (1995) Citrus tristeza virus and its aphid vector Toxoptera citricida Kirkaldy: serious threats to citrus production in the Caribbean Countries, Central America and North America. Plant Dis 79(5):437–445
Roy A, Brlansky RH (2009) Population dynamics of a Florida Citrus tristeza virus isolate and aphid-transmitted sub-isolates: identification of three genotypic groups and recombinants after aphid transmission. Phytopathology 99:1297–1306
Roy A, Brlansky RH (2010) Genome analysis of an orange stem pitting Citrus tristeza virus isolate reveals a novel recombinant genotype. Virus Res 151:118–130
Roy A, Ramachandran P, Brlansky RH (2003) Grouping and comparison of Indian citrus tristeza virus isolates based on coat protein gene sequences and restriction analysis patterns. Arch Virol 148(4):707–722
Roy A, Choudhary N, Hartung JS, Brlansky RH (2013) The prevalence of the Citrus tristeza virus trifoliate resistance breaking genotype among Puerto Rican isolates. Plant Dis 97:1227–1234
Rubio L, Ayllon MA, Kong P, Fernandez A, Polek M, Guerri J, Moreno P, Falk BW (2001) Genetic variation of isolates from California and Spain: evidence for mixed infections and recombination. J Virol 75:8054–8062
Rubio L, Guerri J, Moreno P (2013) Genetic variability and evolutionary dynamics of viruses of the family Closteroviridae. Front Microbiol 4:151
Ruiz-Ruiz S, Moreno P, Guerri J, Ambrós S (2007) A real-time RT-PCR assay for detection and absolute quantitation of Citrus tristeza virus in different plant tissues. J Virol Methods 145:96–105
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning—a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Satyanarayana T, Gowda S, Ayllón MA, Dawson WO (2004) Closterovirus bipolar virion: evidence for initiation of assembly by minor coat protein and its restriction to the genomic RNA 5′ region. Proc Natl Acad Sci USA 101:799–804
Suastika G, Natsuaki T, Terui H, Kano T, Ieki H, Okuda S (2001) Nucleotide sequence of Citrus tristeza virus seeding yellows isolate. J Gen Plant Pathol 67:73–77
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Vives MC, Rubio L, Lopez C, Navas-Castillo J, Albiach-Marti MR, Dawson WO, Guerri J, Flores R, Moreno P (1999) The complete genome sequence of the major component of a mild citrus tristeza virus isolate. J Gen Virol 80(3):811–816
Vives MC, Rubio L, Sambade A, Mirkov TE, Moreno P (2005) Evidence of multiple recombination events between two RNA sequence variants within a Citrus tristeza virus isolate. Virology 331:232–237
Wang J, Bozan O, Kwon S-J, Dang T, Rucker T, Yokomi RK, Lee RF, Folimonova SY, Krueger RR, Bash J, Greer G, Diaz J, Serna R, Vidalakis G (2013) Past and future of a century old Citrus tristeza virus collection: a California citrus Germplasm tale. Front. Microbiol 4:366. https://doi.org/10.3389/fmicb.2013.00366
Weng Z, Barthelson R, Gowda S, Hilf ME, Dawson WO, Galbraith DW, Xiong Z (2007) Persistent infection and promiscuous recombination of multiple genotypes of an RNA virus within a single host generate extensive diversity. PLoS ONE 2(9):E917
Xiao-yun W, Xiao-fei C, Lu L, Xiao-xia W (2012) Genetic diversity and global distribution of Citrus tristeza virus (CTV) strains. J Northeast Agric Univ 19:9–18
Yang ZN, Mathews DM, Dodds JA, Mirkov TE (1999) Molecular characterization of an isolate of Citrus tristeza virus that causes severe symptoms in sweet orange. Virus Genes 19:131–142
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7(1–2):203–214
Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ (eds) Evolving genes and proteins. Academic, New York, pp 97–166
Acknowledgements
Authors acknowledge the laboratory as well as library assistance from BCKV, West Bengal. We further are thankful to the DBT for providing grant (BT/04/NE/2009) under the project Advanced level Institutional Biotech Hub, CHF, CAU, Pasighat.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
42360_2018_56_MOESM1_ESM.jpg
Eco RI digested CP gene insert cloned in pGEM-T Easy Vector. Lane M represents 100 bp DNA ladder (Xcelris genomics, Ahmedabad, India) (JPEG 94 kb)
Rights and permissions
About this article
Cite this article
Singh, A.K., Meetei, N.T., Sutradhar, M. et al. Prevalence of severe Citrus tristeza virus infection in Khasi mandarin throughout the north-eastern states of India: an evidence study based on coat protein gene analysis. Indian Phytopathology 71, 435–443 (2018). https://doi.org/10.1007/s42360-018-0056-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42360-018-0056-5