Abstract
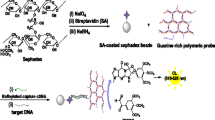
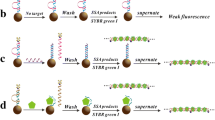
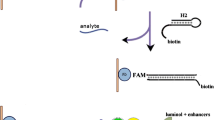
We report on a highly sensitive chemiluminescence (CL) assay for the detection of specific DNA sequence, where streptavidin-modified polystyrene microspheres (PS–SA) coupled with hybridization chain reaction (HCR) were employed as dual amplification platform. Briefly, a “sandwich-type” detection strategy was proposed in our design, which involved capture probe DNA immobilized on the surface of carboxyl-terminated magnetic beads (MB) and the reporter DNA combined with the HCR trigger immobilized on the surface of PS–SA. After that, two hairpin-type monomers were added to initiate the HCR. In the detection system, CL signal was obtained via the instantaneous derivatization reaction between 3,4,5-trimethoxylphenylglyoxal (TMPG) and guanine bases in the target and the HCR complex binding on the magnetic beads. As compared to traditional sandwich type (capture/target/reporter) assays, this dual-amplified strategy for sequence-specific DNA detection showed better specificity, lower detection limit and wider linear response range (linear range of 0.01 fmol–10 pmol and a low detection limit of 5 amol). Moreover, this approach could be easily extended to detect a wide range of specific DNA sequences by modification of the hybridization region. We believe this simple technique will present a significant step towards early diagnoses of diseases.










Similar content being viewed by others
References
Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;70:467–70.
Husale S, Persson HH, Sahin O. DNA nanomechanics allows direct digital detection of complementary DNA and microRNA targets. Nature. 2009;462:1075–8.
Lam B, Das J, Holmes RD, Live L, Sage A, Sargent EH, Kelley SO. Solution-based circuits enable rapid and multiplexed pathogen detection. Nat Commun. 2013;4:2001–8.
Xiao HJ, Hak HC, Kong DM, Shen HX. Sequence-specific detection of nucleic acids utilizing isothermal enrichment of G-quadruplex DNAzymes. Anal Chim Acta. 2012;729:67–72.
Li C, Karadeniz H, Canavar E, Erdem A. Electrochemical sensing of label free DNA hybridization related to breast cancer 1 gene at disposable sensor platforms modified with single walled carbon nanotubes. Electrochim Acta. 2012;82:137–42.
Nascimento JM, Garcia S, Saia-Cereda VM, Santana AG, Brandao-Teles C, Zuccoli GS, Junqueira DG, Reis-de-Oliveira G, Baldasso PA, Cassoli JS, Martins-de-Souza D. Proteomics and molecular tools for unveiling missing links in the biochemical understanding of schizophrenia. Proteom Clin Appl. 2016;10:1148–58.
Ye KH, Manzano M, Muzzi R, Gin KYH, Saeidi N, Goh SG, Tok AIY, Marks RS. Development of a chemiluminescent DNA fibre optic genosensor to Hepatitis A Virus (HAV). Talanta. 2017;174:401–8.
Zou B, Ma Y, Wu H, Zhou G. Signal amplification by rolling circle amplification on universal flaps yielded from target-specific invasive reaction. Analyst. 2012;137:729–34.
Wang X, Lou X, Wang Y, Guo Q, Fang Z, Zhong X, Mao H, Jin Q, Wu L, Zhao H, Zhao J. QDs-DNA nanosensor for the detection of hepatitis B virus DNA and the single-base mutants. Biosens Bioelectron. 2010;25:1934–40.
Chen XH, Roloff A, Seitz O. Consecutive signal amplification for DNA detection based on de novo fluorophore synthesis and host–guest chemistry. Angew Chem Int Ed. 2012;51:4479–83.
Niu S, Jiang Y, Zhang S. Fluorescence detection for DNA using hybridization chain reaction with enzyme-amplification. Chem Commun. 2010;46:3089–91.
Qi YY, Xiu FR, Li BX. One-step homogeneous non-stripping chemiluminescence metal immunoassay based on catalytic activity of gold nanoparticles. Anal Biochem. 2014;449:1–8.
Cai S, Lau CW, Lu JZ. Sequence-specific detection of short-length DNA via template-dependent surface-hybridization events. Anal Chem. 2010;82:7178–84.
Qi Y, Li B. A sensitive, label-free, aptamer-based biosensor using a gold nanoparticle-initiated chemiluminescence system. Chem Eur J. 2011;17:1642–8.
Bi S, Zhao T, Luo B. A graphene oxide platform for the assay of biomolecules based on chemiluminescence resonance energy transfer. Chem Commun. 2012;48:106–8.
Wang HQ, Liu WY, Wu Z, Tang LJ, Xu XM, Yu RQ, Jiang JH. Homogeneous label-free genotyping of single nucleotide polymorphism using ligation-mediated strand displacement amplification with DNAzyme-based chemiluminescence detection. Anal Chem. 2011;83:1883–9.
Li J, Deng T, Chu X, Yang R, Jiang J, Shen G, Yu R. Rolling circle amplification combined with gold nanoparticle aggregates for highly sensitive identification of single-nucleotide polymorphisms. Anal Chem. 2010;82:2811–6.
Liu Y, Wu Z, Zhou G, He Z, Zhou X, Shen A, Hu J. Simple, rapid, homogeneous oligonucleotides colorimetric detection based on non-aggregated gold nanoparticles. Chem Commun. 2012;48:3164–6.
Vezocnik V, Rebolj K, Sitar S, Otaa K, Znidari MT, Strus J, Sepci K, Pahovnik D, Macek P, Zagar E. Size fractionation and size characterization of nanoemulsions of lipid droplets and large unilamellar lipid vesicles by asymmetric-flow field-flow fractionation/multi-angle light scattering and dynamic light scattering. J Chromatogr A. 2015;1418:185–91.
Branch SD, Lines AM, Lynch J, Bello JM, Heineman WR, Bryan SA. Optically transparent thin-film electrode chip for spectroelectrochemical sensing. Anal Chem. 2017;89:7324–32.
Chuang TL, Wei CS, Lee SY, Lin CW. A polycarbonate based surface plasmon resonance sensing cartridge for high sensitivity HBV loop-mediated isothermal amplification. Biosens Bioelectron. 2012;32:89–95.
Zhai J, Cui H, Yang R. DNA based biosensors. Biotechnol Adv. 1997;15:43–58.
Park SJ, Taton TA, Mirkin CA. Array-based electrical detection of DNA with nanoparticle probes. Science. 2002;295:1503–6.
Zhang Y, Pothukuchy A, Shin W, Kim Y, Heller A. Detection of approximately 10(3) copies of DNA by an electrochemical enzyme-amplified sandwich assay with ambient O(2) as the substrate. Anal Chem. 2004;76:4093–7.
Caruana DJ, Heller A. Enzyme-amplified amperometric detection of hybridization and of a single base pair mutation in an18-base oligonucleotide on a 7-mm-diameter microelectrode. J Am Chem Soc. 1999;121:769–74.
Patolsky F, Lichtenstein A, Willner I. Detection of single-base DNA mutations by enzyme-amplified electronic transduction. Nat Biotechnol. 2001;19:253–7.
Shimron S, Wang F, Orbach R, Willner I. Amplified detection of DNA through the enzyme-free autonomous assembly of hemin/G-quadruplex DNAzyme nanowires. Anal Chem. 2012;84:1042–8.
Chai Y, Tian D, Wang W, Cui H. A novel electrochemiluminescence strategy for ultrasensitive DNA assay using luminol functionalized gold nanoparticles multi-labeling and amplification of gold nanoparticles and biotin–streptavidin system. Chem Commun. 2010;46:7560–2.
Zhou X, Xing D, Zhu D, Jia L. Magnetic bead and nanoparticle based electrochemiluminescence amplification assay for direct and sensitive measuring of telomerase activity. Anal Chem. 2009;81:255–61.
Authier L, Grossiord C, Brossier P, Limoges B. Gold nanoparticle-based quantitative electrochemical detection of amplified human cytomegalovirus DNA using disposable microband electrodes. Anal Chem. 2001;73:4450–6.
Dirks RM, Pierce NA. Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci USA. 2004;101:15275–8.
Huang J, Wu Y, Chen Y, Zhu Z, Yang Y, Yang CJ, Wang K, Tan W. Pyrene-excimer probes based on the hybridization chain reaction for the detection of nucleic acids in complex biological fluids. Angew Chem Int Ed. 2011;50:401–4.
Song WQ, Zhu KL, Cao ZJ, Lau CW, Lu JZ. Hybridization chain reaction-based aptameric system for the highly selective and sensitive detection of protein. Analyst. 2012;137:1396–401.
Wang XZ, Jiang AW, Hou T, Li HY, Li F. Enzyme-free and label-free fluorescence aptasensing strategy for highly sensitive detection of protein based on target-triggered hybridization chain reaction amplification. Biosens Bioelectron. 2015;70:324–9.
Hao YL, Guo QQ, Wu HY, Guo LQ, Zhong LS, Wang J, Lin TR, Fu FF, Chen GN. Amplified colorimetric detection of mercuric ions through autonomous assembly of G-quadruplex DNAzyme nanowires. Biosens Bioelectron. 2014;52:261–4.
Kojima E, Ohba Y, Kai M, Ohkura Y. Phenylglyoxal and glyoxal as fluorogenic reagents selective for N-terminal tryptophan-containing peptides. Anal Chim Acta. 1993;280:157–62.
Yu CY, Yin BC, Ye BC. A universal real-time PCR assay for rapid quantification of microRNAs via the enhancement of base-stacking hybridization. Chem Commun. 2013;49:8247–9.
Yin BC, Liu YQ, Ye BC. One-step, multiplexed fluorescence detection of microRNAs based on duplex-specific nuclease signal amplification. J Am Chem Soc. 2012;134:5064–7.
Zuo X, Xia F, Xiao Y, Plaxco KW. Sensitive and selective amplified fluorescence DNA detection based on exonuclease III-aided target recycling. J Am Chem Soc. 2010;132:1816–8.
Shi M, Zheng J, Tan Y, Tan J, Li J, Li Y, Li X, Zhou Z, Yang R. Ultrasensitive detection of single nucleotide polymorphism in human mitochondrial DNA utilizing ion-mediated cascade surface-enhanced Raman spectroscopy amplification. Anal Chem. 2015;87:2734–40.
Ge J, Zhang LL, Liu SJ, Yu RQ, Chu X, Chen A. A highly sensitive target primed rolling circle amplification (TPRCA) method for fluorescent in situ hybridization detection of microRNA in tumor cells. Anal Chem. 2014;86:1808–15.
Deng R, Tang L, Tian Q, Wang Y, Lin L, Li J. Toehold-initiated rolling circle amplification for visualizing individual microRNAs in situ in single cells. Angew Chem Int Ed. 2014;53:2389–93.
Cao ZJ, Peng QW, Qiu X, Liu CY, Lu JZ. Highly sensitive chemiluminescence technology for protein detection using aptamer-based rolling circle amplification platform. J Pharceutical Anal. 2011;1:159–65.
Wang X, Lau CW, Kai M, Lu JZ. Hybridization chain reaction-based instantaneous derivatization technology for chemiluminescence detection of specific DNA sequences. Analyst. 2013;138:2691–7.
Cai S, Cao ZJ, Lau CW, Lu JZ. Label-free technology for the amplified detection of microRNA based on the allosteric hairpin DNA switch and hybridization chain reaction. Analyst. 2014;139:6022–7.
Su J, Zhang HJ, Jiang BY, Zheng HZ, Chai YQ, Yuan R, Xiang Y. Dual signal amplification for highly sensitive electrochemical detection of uropathogens via enzyme-based catalytic target recycling. Biosens Bioelectron. 2011;29:184–8.
Takalkar S, Baryeh K, Liu GD. Fluorescent carbon nanoparticle-based lateral flow biosensor for ultrasensitive detection of DNA. Biosens Bioelectron. 2017;98:147–54.
Zhong D, Yang KC, Wang YY, Yang XM. Dual-channel sensing strategy based on gold nanoparticles cooperating with carbon dots and hairpin structure for assaying RNA and DNA. Talanta. 2017;175:217–23.
Peng QW, Cao ZJ, Lau CW, Kai M, Lu JZ. Aptamer-barcode based immunoassay for the instantaneous derivatization chemiluminescence detection of IgE coupled to magnetic beads. Analyst. 2011;136:140–7.
Tang W, Wang D, Xu Y, Li N, Liu F. A self-assembled DNA nanostructure-amplified quartz crystal microbalance with dissipation biosensing platform for nucleic acids. Chem Commun. 2012;48:6678–80.
Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–81.
Taton TA, Lu G, Mirkin CA. Two-color labeling of oligonucleotide arrays via size-selective scattering of nanoparticle probes. J Am Chem Soc. 2001;123:5164–5.
Nam JM, Stoeva SI, Mirkin CA. Bio-bar-code-based DNA detection with PCR-like sensitivity. J Am Chem Soc. 2004;126:5932–3.
Feng X, Luoa XT, Hsing IM. Sensitive immobilization-free electrochemical DNA sensor based on isothermal circular strand displacement polymerization reaction. Biosens Bioelectron. 2012;35:230–4.
Gao FL, Lei JP, Ju HX. Label-free surface-enhanced Raman spectroscopy for sensitive DNA detection by DNA-mediated silver nanoparticle growth. Anal Chem. 2013;85:11788–93.
Zhang ZP, Zhou JY, Tang AA, Wu ZY, Shen GL, Yu RQ. Scanning electrochemical microscopy assay of DNA based on hairpin probe and enzymatic amplification biosensor. Biosens Bioelectron. 2010;25:1953–7.
Zhao C, Wu L, Ren J, Qu X. A label-free fluorescent turn-on enzymatic amplification assay for DNA detection using ligand-responsive G-quadruplex formation. Chem Commun. 2011;47:5461–3.
Mei Z, Tang L. Surface-plasmon-coupled fluorescence enhancement based on ordered gold nanorod array biochip for ultrasensitive DNA analysis. Anal Chem. 2017;89:633–9.
Alonso-Cristobal P, Vilela P, El-Sagheer A, Lopez-Cabarcos E, Brown T, Muskens OL, Rubio-Retama J, Kanaras AG. Highly sensitive DNA sensor based on upconversion nanoparticles and graphene oxide. ACS Appl Mater Interfaces. 2015;7:12422–9.
Acknowledgements
This work was supported by the Natural Science Foundation of China (Nos. 21375025 and 21675030).
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Zhou, Y., Wang, Y., Wang, X. et al. Polystyrene Microspheres Coupled with Hybridization Chain Reaction for Dual-Amplified Chemiluminescence Detection of Specific DNA Sequences. J. Anal. Test. 1, 306–314 (2017). https://doi.org/10.1007/s41664-017-0042-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41664-017-0042-4