Abstract
Thousands of results of scientific publications on the topic of COVID-19 have emerged within a short time, raising significant challenges for scholars to organize the research, or even synthesize the knowledge in a timely or comprehensive manner. To facilitate this, we have developed a geospatial and semantic mapping platform to search and organize these large and unmapped digital collections. The semantic map visualizes research topics based on customized natural language processing algorithms, which helps users to identify their content of specific interest beyond keyword searches in web search engines. The resultant geospatial map extracts all the location names mentioned in the publications, illustrating where COVID-19 studies have been conducted and where neglected study areas might exist. The system has been deployed as a web service for public access.
Similar content being viewed by others
Introduction
In December 2019, Chinese authorities identified an outbreak of a new coronavirus, SARS-CoV-2. This virus can infect people of all ages and cause mild to severe illness known as coronavirus disease 2019 (COVID-19). The COVID-19 pandemic has been causing unprecedented consequences in human society (Kapitsinis 2020; Zhang et al. 2020). It has infected more than 50 million people and caused the deaths of over 1.2 million people as of November 9, 2020.Footnote 1 Many efforts, including social distancing, wearing face masks, city/region lockdowns, and contact tracing and quarantine, have been adopted to slow and stop the spread of the life-threatening virus (Huang et al. 2020). Society has staggered under the significant impact of the pandemic on everyday life and has been waiting for the pandemic to end and for a cure or vaccine (Hu et al. 2020).
The research community has also been actively reacting to the sustained risk of the global spread of COVID-19. Freely available digital collections containing a large quantity of COVID-19 scientific publications have been generated and are still expanding. A loose-knit army of scientists, ranging from public health, biology, social sciences, to computational science, have been devoted to studying the pandemic and its ecosystems according to their respective expertise. The explosive volumes of research in this area, however, are challenging users’ ability to seek the most relevant or valuable studies. As Brainard (2020) estimated, more than 23,000 papers have been published between January and early May of 2020, and the publication numbers double every 20 days. This adds to the difficulty in weeding through the sheer volume of work to find the most valuable and relevant work.
This has created the need for tools that will help researchers organized, so they can push forward research frontiers without extraneous time spent repeating searches. From the geospatial perspective, it is notable that a global effort is committing to research, but it is also noted that existing studies are unevenly distributed, potentially due to the underlying socioeconomic inequality and political topography. Searching through COVID-19 papers using spatial information is especially tedious, because geographic information is normally mixed in with other contexts, such as authors’ addresses, affiliations, conference locations, and more importantly the study areas. Most general search engines do not provide geospatial maps indicating the spread of scientific publications. Therefore, a more advanced search platform is required.
In this paper, we develop a knowledge discovery system specifically for COVID-19 scientific publications. Differentiated from most general academic search engines (e.g., Google Scholar, Semantic Web, and Scopus), the system provides both semantic and geospatial maps for COVID-19 research papers. It highlights the following aspects:
-
1.
Multi-faceted search options: Our system can not only search the body texts of COVID-19 publications but also search the titles, abstracts, tables, and figures in these publications. The multi-faceted search options provide users full flexibility to investigate different components of scientific publications and help users to target more relevant information.
-
2.
Semantic map: Our system presents a semantic map to visualize the topics in COVID-19 research publications. Advanced natural language processing (NLP) algorithms are integrated into this semantic map for flexible topic discovery, which users can customize by specifying different NLP algorithms and parameters.
-
3.
Geospatial map: The location names mentioned in COVID-19 publications are extracted and visualized on a geospatial map, illustrating where COVID-19 studies have been conducted and where neglected study areas might exist.
The demo website is deployed at http://covid19papers.njit.edu/. All the codes for this open-source system are shared on GitHub (https://github.com/cordsearch/cord-search.github.io), so users can change and deploy our website to their local environment.
Related Work
General search engines are intuitive and easily accessible tools for the general public to search for information. In response to COVID-19-related queries, general search engines, such as Google and Bing, can return a variety of resources and personalized search results, which include government guidelines, news, and other multi-media content (Makhortykh et al. 2020). However, articles containing misinformation and disinformation might exist in the search results, which could cause confusion and pose threat to public health in this pandemic (Jayasinghe et al. 2020). Academic search engines that focus on scientific publications (e.g., Google Scholar) could return more reliable information on COVID-19. These academic search engines tend to use keyword co-appearance and other semantic technologies to search for relevant literature (Fricke 2018), but they cannot extract meaningful patterns from findings across COVID-19 publications. This clearly indicates that any search platform that is developed should have advanced filtering abilities.
COVID-19 research literature has become freely available to the global research community and has prompted the development of a new search engine using the rich content text of these publications. The large open-access dataset (Allen Institute For AI 2020) has allowed researchers to apply cutting-edge NLP and other artificial intelligence (AI) techniques to generate knowledge to support the ongoing fight against infectious disease. There is an increasing need for these approaches because it is challenging for researchers and decision-makers to keep up with the quickly growing COVID-19 literature. One data provider, Kaggle,Footnote 2 has been providing several notebooks and discussions related to search strategies for the massive body of COVID-19 scientific publications. However, keyword-based search systems only consider titles and abstracts of publications in their search algorithms (Dai and Callan 2019). Other search systems may dig deeper to include the publication content and use advanced semantic technologies to construct the knowledge graph of the literature (Wise et al. 2020). But, such systems usually do not allow users to customize search strategies (models and parameters) and directly return results based on a predefined search algorithm. Although systems using analytical search strategies can answer predefined questions accurately, what is lacking is the generality to answer other questions (Su et al. 2020).
There are many ways to organize information in the literature. The most straightforward way would be reading all relevant articles and summarize the main ideas manually (Biljecki 2016). However, this approach makes it impossible to obtain a large number of results in a timely manner. The semantic map can automatically assign papers to topic clusters in a two-dimensional visualization reference system (Ahearn et al. 2013). Standard algorithms have also been applied to cluster the COVID-19 literature in some search systems (Solovyev et al. 2020). These clustering algorithms use a predefined clustering model and a fixed number of clusters, which do not always generate meaningful or consistent results when the search content is varied. And, while each of the algorithms described has its individual advantages and disadvantages, the provision of a single clustering algorithm is insufficient to present a holistic view of the publication topics. Additionally, the parameters of each algorithm should have the capacity for a variation to ensure flexibility. The incorporation of different and customizable algorithms into our system ensures flexibility to help users explore and retrieve more meaningful results.
System Design
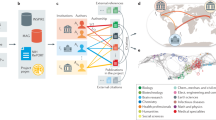
Our system uses state-of-the-art language modeling methods to analyze publication content and dynamically visualizes the results. Users can customize the analyses and visualizations by specifying different models and parameters. Figure 1 shows the workflow of the geospatial and semantic mapping platform. First, the original COVID-19 research papers are pre-processed; then, the text data is embedded in high-dimensional space. Next, different dimension reduction algorithms are used to generate two-dimensional topic coordinates and extract geographic locations from each paper. Finally, these coordinates are presented as visualizations on a semantic map and a geospatial map. This section will introduce the system design in detail.
Data Preparation
First, we crawled through all the data from CORD-19-research-challenge using Kaggle (Allen Institute For AI 2020), which consists of the publications from different sources, such as Chan Zuckerberg Initiative, PubMed Central, BioRxiv, and MedRxiv. The dataset contained over 341,713 scholarly articles about COVID-19, SARS-CoV-2, and related coronaviruses (as of November 8, 2020). Among these publications, 222,600 had full text, 243,737 had abstracts, and 117,596 articles were published in 2020. The papers in PDF format were converted to pure text using pdfMiner (Shinyama 2015) and optical character recognition programs (Smith 2007). The data was further pre-processed by removing the null values and non-alphabet texts.
Embedding
The process of embedding means converting high-dimensional text data to low-dimensional vectors. It is a necessary step before applying NLP algorithms, because most NLP algorithms expect inputs to be numerical feature vectors with a fixed size rather than unstructured text data with variable lengths. Different embedding methods have their own advantages and disadvantages in representing the text meanings, and thus, three different embedding methods are used in parallel in the system, thereby creating a system skewing to be more advantageous and flexible.
Bag of Words
The bag of words (BoW) model is a dictionary-based representation used in NLP and information retrieval (Dai and Callan 2019). It represents documents based on the frequencies of word occurrences.
For example: if we have a text corpus with four documents as follows:
-
“This is the first document.”
-
“This is the second second document.”
-
“And the third one.”
-
“Is this the first document?”
Then, we can build a 9-element dictionary (vocab):
[“and”, “document”, “first”, “is”, “one”,“second”, “the”, “third”, “this”]
Each sentence can be embedded to a 9-element vector by counting the occurrences of each corresponding word in the sentence. If a sentence does not have any words included in the dictionary, it will generate a vector of all zeros. Therefore, the text corpus would be represented as
array([[0, 1, 1, 1, 0, 0, 1, 0, 1],
[0, 1, 0, 1, 0, 2, 1, 0, 1],
[1, 0, 0, 0, 1, 0, 1, 1, 0],
[0, 1, 1, 1, 0, 0, 1, 0, 1]]...)
Term frequency-inverse document frequency (tfidf) regularization is usually applied to BoW to reflect the importance of a word to a document in a collection or corpus.
in which tf refers to the number of times a term t occurs in a given document d. The idf component is computed as
where n is the total number of documents in the document set and df(t) is the number of documents in the document set that contain term t.
Word Vectors
The second embedding method leverages pre-trained word vectors to mine the research publications. Instead of treating a word as “existing or not” (0 or 1) as in the BoW method, word vectors contain the semantic meaning of each word by learning from a large text corpus (Bojanowski et al. 2017).
In our implementation, we use the Global Vectors for Word Representation (GLOVE) (Pennington et al. 2014) to embed words and generate the embedding of a document using average pooling. The GLOVE is an unsupervised learning algorithm for obtaining vector representations for words. With GLOVE, we can use pre-trained word embedding to represent words. The training is carried out on the aggregated global word-word co-occurrence statistics derived from a corpus, and the resulting representations showcase interesting linear substructures of the word vector space. Then, we can combine the meaning of each word in an article to represent the meaning of the entire article.
BERT
The Bidirectional Encoder Representations from Transformers (BERT) (Devlin et al. 2018) is a pre-trained language model developed by Google, which can provide context-aware embedding for natural languages. It is a bidirectional transformer pre-trained using a combination of masked language modeling objectives and next sentence predictions on a large corpus comprising the Toronto Book Corpus and Wikipedia. The BERT is conceptually simple and empirically powerful. The pre-trained BERT model can be fine-tuned for new NLP tasks to create state-of-the-art models, such as question answering and language inference, without substantial task-specific architecture modifications.
The BERT model can better represent the semantic meaning in sentences because it can learn context from the given data. We fine-tune the BERT-large-uncased-whole-word-masking (Devlin et al. 2018) version of the BERT model using the COVID-19 publication collection. Due to BERT’s maximum 512 word limitation, we could only use each paper’s title and the first sentence of the abstract for embedding.
All of these embedding methods are provided in the semantic maps of the system. Detailed instructions are introduced in the “Semantic Map” section.
Dimension Reduction
Although COVID-19 publications can be represented as n-dimensional vectors, it is not intuitive to analyze their characteristics and relationships. Visualization can help users compare those embedding vectors intuitively. However, the high-dimensional vectors cannot be directly visualized on a two-dimensional computer screen. In our system, dimension reduction is applied to high-dimensional embedding vectors to generate a two-dimensional coordinate for each publication. This allows for the publications to be displayed on a flat semantic map.
For BoW embedding, we use the latent Dirichlet allocation (LDA) (Blei et al. 2003) to discover abstract topics from a collection of documents. This model is based on the frequency of each word in the whole dataset. The LDA is an unsupervised machine learning model that is used in a variety of domain topic modeling (Pedregosa et al. 2011). It also identifies keywords of a cluster based on how distinctive a word is from others in the cluster.
For GLOVE embedding, we first use the mean embedding vector for all words in a paper to represent the paper. Then, we convert the embedding vectors of the papers to two-dimensional vectors using Principal Component Analysis (PCA) (Halko et al. 2011). Next, the K-means (Sculley 2010) algorithm is used to classify these papers. Finally, in each cluster, we calculate the most frequently used words as the cluster’s keywords.
For BERT embedding, the t-sne (Maaten and Hinton 2008) has a better dimension reduction performance than the LDA and PCA (McInnes et al. 2020). The t-sne converts similarities between data points to joint probabilities and simultaneously tries to minimize the Kullback-Leibler divergence between the joint probabilities of the low-dimensional embeddings and the high-dimensional data. We use t-sne to cluster COVID-19 publications and extract keywords based on word frequencies. Table 1 shows an example that identifies five topic clusters using this approach. Topic 1 is about virus infection; topic 2 includes clinic case studies; topic 3 is the protein level studies; topic 4 talks about the general virus; and topic 5 is about the distribution of the influence. Users can also change the number of topic clusters easily by specifying a cluster quantity in the user interface (UI).
Multi-faceted Search
We break each COVID-19 research paper into multiple components (title, abstract, paragraphs, tables, and figures) and index them with the Elasticsearch algorithm (Gormley and Tong 2015). The figures and tables are also illustrative components in research papers besides the main texts and abstracts (Wong 2010); hence, we extract the table and figure names from the papers for searching. Then, we use Elasticsearch with the BM25 (Robertson and Zaragoza 2009) method as the back-end service to search the relevant documents. We develop middleware to covert the results in JSON format returned by the Elasticsearch, into a more readable version and display the paragraphs in search results one by one. The user-friendly search experience of the system is similar to the one allowed by the Google search engine. We hope in this way users can quickly get started with the system.
For all the text fields, the BM25 algorithm is used to query and rank results in the system. The BM25 is a classic information retrieval algorithm based on the BoW representation. It is the default algorithm in Apache Lucene (also in Solr and Elastic search) (Robertson and Zaragoza 2009). We calculate the BM25 as:
where q is the query, tq is the word’s frequency in a query, |d| is the length of document d in words, and avgdl is the average document length in the corpus. k1 and b are hyperparameters, which are set as 2 and 0.75 accordingly based on suggestions in the literature (Robertson and Zaragoza 2009).
Spatial Information Analysis
There is much structured spatial information related to publications such as author address, institution location, and conference location. However, readers would likely care more about the case locations of these studies than other spatial contexts. Although case studies in COVID-19 research publications contain such spatial information, there are no explicit coordinates provided. Therefore, the spatial information needs to be extracted from the unstructured text and then geocoded. This task is usually referred to as geoparsing. We build a 3-step geoparsing pipeline as follows.
Place Name Extraction
The first step is to extract text that contains spatial information. We use the name entity recognition (NER) tool SpaCy.Footnote 3 SpaCy is a natural language processing library based on convolutional neural networks. It is trained on large amounts of open datasets and has a high precision for NER tasks. The SpaCy is implemented heavily in C-level data structure and functions. The C programming language, in its core, guarantees the high performance of the SpaCy, so it can efficiently deal with a large number of publications in our system.
The SpaCy NER tool can identify PERSON, NORP (e.g., nationalities and political groups), FACILITY (e.g., buildings, airports, and highways), ORG (e.g., companies, agencies, and institutions), GPE (e.g., countries, cities, and states), LOC (e.g., non-GPE locations, mountain ranges, and bodies of water), and other types of entities. Given our interest in spatial information, we only keep FACILITY, ORG, GPE, and LOC in the extracted entities.
Location Disambiguation
Non-place names could be recognized as place names in the NER. We call these false-positive cases. This happens because natural language, even in research papers, is highly variable. Place names may appear in ways that are not expected. Rules like place names following prepositions “in” or “at” will not always be applicable. Therefore, the NER tools tolerate many irregularities, which result in false positives.
Since COVID-19 studies are conducted all over the world, identifying the country name(s) of a study is the fundamental need when we extract spatial information. We develop a language-agnostic architecture that uses neural networks to infer the correct country from a set of locations in a piece of text. The input is the word2vec (Mikolov et al. 2013) vector of a place name, and the output is the probability of a match between the place name and an entry in the Gazetteer. The training data is a newly annotated dataset labeled with Prodigy.Footnote 4 As shown in Fig. 2, we use three dense layers in the neural network. Each layer has 512 hidden dimensions and follows the Relu activation function with a dropout rate of 0.5. Finally, we obtain the probability of a match as a confidence score. If the probability is greater than 0.5, we consider it as a correct match.
Gazetteer
Because of the large amount of publication data available, it is too time-consuming to use online gazetteer services for geocoding. We choose to build a local gazetteer, using the data from Geonames.Footnote 5 Geonames is a downloadable geographical database covering all countries with over 11 million place names. We index the database in Elasticsearch.Footnote 6 Elasticsearch is an in-memory, distributed, RESTful search engine. The key feature of Elasticsearch is that it keeps all the data in the memory. In most cases, input/output (I/O) to the memory is dozens of times faster than I/O to the hard disk. Therefore, without the need for frequent hard disk I/O, Elasticsearch can retrieve the gazetteer much faster than other methods.
Visualization
Our system includes a semantic map and a geospatial map for visualization. The two maps allow users to investigate COVID-19 research publications through two different perspectives.
Semantic Map
Each publication is represented as a point on the semantic map (Fig. 3). The color of a point indicates to which topic the publication belongs. Users can choose different algorithms and cluster numbers to generate different styles of the semantic map using the two drop-down boxes on top of the semantic map. In the left box, users can select one of the models introduced in the “Embedding” and “Dimension Reduction” sections, and in the right box, users can specify an arbitrary number of topic clusters. Then, the semantic map will be computed and rendered according to these selections. The distance between the two points represents the semantic similarities between the two publications. The more related two publications are, the closer their representing points are in the semantic map. The keywords of each category are shown as a list below the map (Fig. 3). Users can type in the search bar and check a paper component (abstract, body text, paragraph, or figure and table) to create a customized query for COVID-19 research publications (Fig. 3). The system will retrieve relevant publications based on the query and display the search results in the result field as shown in Fig. 4. The highlighted phrases are the matched key phrases between the query and papers. For example, if we search “New York” by checking the abstract search mode, our system will retrieve all articles with “New York” in their abstracts and highlight the key phrases.
The semantic map and the search function are correlated. Users can view an article by using the mouse to hover over any marked point on the semantic map, as well as the topic of the paper. Clicking on the point fills information into the search bar, and a search is conducted by the system to retrieve the relevant publications, which are also highlighted in the result field. Users can further filter search results by selecting a topic category. For example, users can choose topic cluster c-1 to only retrieve articles in that category (Fig. 5) or use text to filter the results. As shown in Fig. 6, after searching for “New York” in all publications, we type “SEIR” into the filter box. The system further filters the publications from the previous search results and finally gets one publication containing both “New York” and “SEIR.”
The Web UI of the system is created using Vanilla JS and HTML. The operations on the Web UI are designed to be user-friendly and the system is also designed to be mobile-device friendly. The Web UI also has the ability to respond with real-time search results, even while typing the search words into the search bar.
Geospatial Map
The geospatial map displays the spatial information mentioned in COVID-19 research publications. If multiple occurrences of the same location appear in the same publication, they are considered as duplicate locations and are only counted once. Thus, users can easily identify the hotspots in COVID-19 research by examining the spatial view of all the publications. At the same time, the “blank” spaces in the map can indicate neglected study areas where little research has been conducted.
The web interface uses Openlayers framework to visualize the extracted spatial information. The interface provides an aggregated view of the publications, where the circle size is proportional to the number of publications related to the given location (Figs. 7 and 8). When a circle is clicked, a list of related publications pops up in the map display. Users can use the mouse or touchpad to zoom in to navigate the cluster details. Zooming tools are also provided in the left side corner to help users navigate the map.
Discussion
The semantic and geospatial mapping platform described previously has been designed for researchers and decision-makers to keep pace with and explore the growing torrent of new scientific publications related to COVID-19. The uniqueness of our semantic map lies in its ability to be customized. We integrate a variety of natural language processing models into our system and enable users to specify the parameters for fine-scale topic classifications. The combination of models, each of which has its advantages and disadvantages if separated, provides researchers a holistic view of scientific publications related to COVID-19. While most other systems only provide a single search approach with fixed predefined parameters, our system provides a collection of search algorithms with flexible parameters for users to analyze COVID-19 research publications. For example, users can choose the model as well as the number of topic clusters to categorize the publications. When users zoom in to check research publications in a specific topic category, they can not only easily identify related papers of that topic but can also navigate to the borderline of topic clusters to identify the related studies.
The highlight of our geospatial map is its focus on mapping location names mentioned in research publications. There are plenty of COVID-19 dashboards available for visualizing the locations of reported cases or related news articles. However, our geospatial map only extracts all the location names mentioned by the research community in publications, offering users a chance to look into the spatial distribution of COVID-19 research. The geospatial map can clearly illustrate where COVID-19 studies have been conducted and where neglected study areas might exist. The knowledge gaps across locations could possibly reveal the unevenness of resource allocations, given the similar incidence rates of COVID-19.
The system is fast and accurate in searching through the different components of research publications. It is hosted on a dedicated server with an eight-core CPU and 32GB RAM. When opening the website for the first time, the system may take a few seconds to load the semantic map. This is the only delay a user should encounter. After loading the page, the system responds to users’ actions with almost no delay. For example, when a user enters a keyword into the search bar, the system will return the search results in near real time. The system can accurately retrieve related publications based on keywords entered by users.
There are some limitations in the search and the semantic and geospatial maps. First, because all parts of the system implement well-developed algorithms from previous literature, we have not conducted separate evaluations on the accuracy of each algorithm. Second, the research publication dataset is from an open-access platform (Kaggle). Many recent publications have only undergone a fast-tracked review process because publishers want to catch up with the trend, which might result in uneven quality in terms of some research publications. This may skew the search a little but is out of our control. In future studies, we would like to further investigate the quality of research publications, e.g., by ranking the publications based on their number of citations. In addition, it is acknowledged that the geospatial map might not exactly reflect the study areas, because the geocoding process used in this study might only achieve 83% accuracy (Halterman 2019). Some location names might be geocoded incorrectly or cannot be geocoded. This requires further research and refinement of the geocoding tools to improve accuracy. We hope to further improve the accuracy of the location extraction process by curating the data and leveraging more advanced machine learning algorithms. Third, although new research is published every day continuously, we can only update website data weekly due to the limited computing power and resources we have. Future research may consider exploring automatic updating methods for the publication dataset to reflect the publication changes in a timelier manner. Finally, the semantic map and geospatial map are currently on different web pages with hyperlinks for users to navigate between the two. This is somewhat intentional, because the two maps do not directly interact with each other. In our view, it is more valuable and easier for users to view one map at a time rather than navigate several maps together on a single web page. The spatial information of the publications can be further analyzed and visualized in future versions of the system. For example, the geospatial maps can show the co-occurrences of study locations in the same publication. Future studies may also focus more on aggregating information from multiple academic databases.
Conclusion
This paper presents the development of a web-based system to store, search, and analyze research papers involving COVID-19. The system includes a semantic map to visualize the semantic topics among COVID-19 studies. The semantic map in the center of the UI utilizes the most advanced deep learning models for flexible topic discovery and visualization, for users to easily find relevant publications, based on desire parameters. The semantic map also provides multi-faceted search functions, through which a user can search the abstracts, body texts, and even figures and tables in those publications. The figure and table search is a unique feature of our system. The system also features a geospatial map to visualize the location names mentioned within the publications. The geospatial map allows for the visualization of research hotspots, as well as the identification of research deserts.
In summary, we believe this new COVID-19 scientific publication exploration system can act as the foundation for a more general platform to contrast knowledge artifacts, generate hypotheses, compare algorithms, and conduct exploratory research. Thus, the system has the potential to evolve into the centerpiece of a cyberinfrastructure ecosystem for COVID-19 knowledge discovery in specific and convergence research in general.
References
Ahearn SC, Icke I, Datta R, DeMers MN, Plewe B, Skupin A (2013) Re-engineering the GIS&T body of knowledge. Int J Geogr Inf Sci 27(11):2227–2245. https://doi.org/10.1080/13658816.2013.802324
Allen Institute For AI (2020) Covid-19 open research dataset challenge (CORD-19). https://www.kaggle.com/allen-institute-for-ai/CORD-19-research-challenge
Biljecki F (2016) A scientometric analysis of selected GIScience journals. Int J Geogr Inf Sci 30(7):1302–1335. https://doi.org/10.1080/13658816.2015.1130831
Blei DM, Ng AY, Jordan MI (2003) Latent dirichlet allocation. J Mach Learn Res 3(Jan):993–1022
Bojanowski P, Grave E, Joulin A, Mikolov T (2017) Enriching word vectors with subword information. Trans Assoc Comput Linguist 5:135–146. https://doi.org/10.1162/tacl.a.00051
Brainard J (2020) Scientists are drowning in Covid-19 papers. Can new tools keep them afloat? Science. https://doi.org/10.1126/science.abc7839
Dai Z, Callan J (2019) Deeper text understanding for IR with contextual neural language modeling. In Proceedings of the 42nd International ACM SIGIR Conference on Research and Development in Information Retrieval, pages 985–988
Devlin J, Chang MW, Lee K, Toutanova K (2018) BERT: pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805
Fricke SN (2018) Semantic scholar. J Med Lib Assoc 106:145–147
Gormley C, Tong Z (2015) Elasticsearch: the definitive guide: a distributed real-time search and analytics engine. O’Reilly Media, Inc
Halko N, Martinsson PG, Tropp JA (2011) Finding structure with randomness: probabilistic algorithms for constructing approximate matrix decompositions. Soc Ind Appl Math Rev 53(2):217–288. ISSN 1095-7200. https://doi.org/10.1137/090771806
Halterman A (2019) Geolocating political events in text. In Proceedings of the Third Workshop on Natural Language Processing and Computational Social Science, NAACL, pages 29–39
Hu T, Yue H, Wang C, She B, Ye X, Liu R, Zhu X, Bao S (2020) Racial segregation, testing sites access, and COVID-19 incidence rate in Massachusetts, USA. Int J Environ Res Public Health 17(24):9528. https://doi.org/10.3390/ijerph17249528
Huang X, Li Z, Jiang Y, Ye X, Deng C, Zhang J, Li X (2020) The characteristics of multi-source mobility datasets and how they reveal the luxury nature of social distancing in the us during the COVID-19 pandemic. medRxiv. https://doi.org/10.1101/2020.07.31.20143016
Jayasinghe R, Ranasinghe S, Jayarajah U, Seneviratne S (2020) Quality of online information for the general public on COVID-19. Patient Educ Couns 103(12):2594–2597
Kapitsinis N (2020) The underlying factors of the COVID-19 spatially uneven spread. Initial evidence from regions in nine EU countries. Reg Sci Policy Pract 12:1027–1045
Maaten LVD, Hinton G (2008) Visualizing data using t-SNE. J Mach Learn Res 9(Nov):2579–2605
Makhortykh M and Urman A and Ulloa Roberto (2020) How search engines disseminate information about COVID-19 and why they should do better. The Harvard Kennedy School (HKS) Misinformation Review, 1. https://misinforeview.hks.harvard.edu/article/how-search-engines-disseminate-information-about-covid-19-and-why-they-should-do-better/
McInnes L, Healy J, Melville J (2020) UMAP: uniform manifold approximation and projection for dimension reduction. arXiv:1802.03426v3
Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J (2013) Distributed representations of words and phrases and their compositionality. In Advances in Neural Information Processing Systems, pages 3111–3119
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830
Pennington J, Socher R, Manning CD (2014) GloVe: Global vectors for word representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), pages 1532–1543
Robertson S, Zaragoza H (2009) The probabilistic relevance framework: BM25 and beyond. Found Trends Inf Retr 3(4):333–389. https://doi.org/10.1561/1500000019
Sculley D (2010) Web-scale k-means clustering. In Proceedings of the 19th International Conference on World Wide Web, pages 1177–1178
Shinyama Y (2015) Pdfminer: Python pdf parser and analyzer. https://pypi.org/project/pdfminer/
Smith R (2007) An overview of the Tesseract OCR engine. In Ninth International Conference on Document Analysis and Recognition (ICDAR 2007), pages 629–633. https://doi.org/10.1109/ICDAR.2007.4376991
Solovyev EM, Nicholas N, Raff C (2020) COVID-19 literature clustering. In Kaggle. https://github.com/MaksimEkin/COVID19-Literature-Clustering. Malware Research Group
Su D, Xu Y, Yu T, Siddique FB, Barezi EJ, Fung P (2020) CAIRE-COVID: a question answering and multi-document summarization system for COVID-19 research. arXiv preprint:arXiv:2005.03975v3
Wise C, Ioannidis VN, Calvo MR, Song X, Price G, Kulkarni N, Brand R, Bhatia P, Karypis G (2020) COVID-19 knowledge graph: accelerating information retrieval and discovery for scientific literature. arXiv preprint:arXiv:2007.12731
Wong B (2010) Design of data figures. Nat Methods 7(665). https://doi.org/10.1038/nmeth0910-665
Zhang X, Ji Z, Zheng Y, Ye X, Li D (2020) Evaluating the effect of city lock-down on controlling COVID-19 propagation through deep learning and network science models. Cities 107:102869
Acknowledgements
This material is partially based upon work supported by the National Science Foundation under Grant Nos. 1739491 and 1937908. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author and do not necessarily reflect the views of the National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This paper is compliant with ethical standards.
Conflict of Interest
The authors declare that they have no conflicts of interest.
Ethical Approval
Not applicable. This study only uses scientific literature published on the topic of COVID-19. For this type of study ethical approval is not required.
Informed Consent
Not applicable. This study only uses scientific literature published on the topic of COVID-19. For this type of study informed consent is not required.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ye, X., Du, J., Gong, X. et al. Geospatial and Semantic Mapping Platform for Massive COVID-19 Scientific Publication Search. J geovis spat anal 5, 5 (2021). https://doi.org/10.1007/s41651-021-00073-y
Accepted:
Published:
DOI: https://doi.org/10.1007/s41651-021-00073-y