Abstract
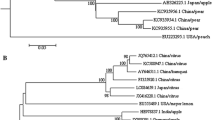
Apple chlorotic leaf spot virus (ACLSV) is a most common pathogen of apples in the world. The occurrence, genetic diversity, recombination patterns and mutation of ACLSV CP in apple trees 80–120 years old from old homesteads and abandoned orchards in Lithuania were investigated. Using RT-PCR, 73.8% of samples tested positive for ACLSV. Specific PCR products to virus CP gene were sequenced and analysed from 39 RNA samples of apple trees collected in different geographic locations. Heterogenic infection of ACLSV in the same apple tree was detected in 11 samples. Fifty Lithuanian sequences of ACLSV with diversity in CP gene were submitted in NCBI and compared to 28 analogous sequences by other authors. The identity of virus CP gene among Lithuanian isolates ranged 81.6–99.7% at the nucleic and 88.6–100% at the amino acids level. ACLSV isolates significantly distinguished into two clusters and seven subclusters on phylogenetic tree. Isolates related to P205, A4, B6 and GC10a were identified in Lithuania, and relationship with Mo-5 and RC was not found. Two recombinant Lithuanian isolates of ACLSV distinguished to separate subcluster R-LT. Lithuanian isolates were characterized by combination of amino acids in conservative sites. The unique single or multiple mutations in CP gene among Lithuanian isolates were observed, and substitution in conservative sites 59, 75 and 79 was also identified. The high prevalence and diversity of latent infection in old apple trees was frequently determined due to the genomic mutation and rarely due to the recombination.

Similar content being viewed by others
References
Adams MJ, Candresse T, Hammond J, Kreuze JF, Martelli GP, Namba S, Pearson MN, Ryu KH, Saldarelli P, Yoshikawa N (2012) Family Betaflexiviride. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Academic Press, London, pp 920–941
Al Rwahnih M, Turturo C, Minafra A, Saldarelli P, Myrta A, Pallas V, Savino V (2004) Molecular variability of Apple chlorotic leaf spot virus in different hosts and geographical regions. J Plant Pathol 86:117–122
Anisimova M, Gascuel O (2006) Approximate likelihood ratio test for branches: a fast, accurate and powerful alternative. Syst Biol 55:539–552
Callaway A, Giesman-Cookmeyer D, Gillock ET, Sit TL, Lommel SA (2001) The multifunctional capsid proteins of plant RNA viruses. Annu Rev Phytopathol 39:419–460
Candresse T, Lanneau M, Rever F, Grasseau N, Macquaire G, German S, Malinowski T, Dunez J (1995) An immunocapture PCR assay adapted to the detection and the analysis of the molecular variability of the Apple chlorotic leaf spot virus. Acta Hortic 386:136–147
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Chen S, Zhou Y, Ye T, Hao L, Guo L, Fan Z, Li S, Zhou T (2014) Genetic variation analysis of apple chlorotic leaf spot virus coat protein reveals a new phylogenetic type and two recombinants in China. Arch Virol 159:1431–1438
Chevenet F, Brun C, Banuls AL, Jacq B, Chisten R (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinform 7:439
Dereeper A, Audic S, Claverie JM, Blanc G (2010) BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 10:8
Desvignes JC, Boye R (1989) Different diseases caused by the chlorotic leafspot virus on the fruit trees. Acta Hortic 235:31–38
Dhir S, Zaidi AA, Hallan V (2013) Molecular characterization and recombination analysis of the complete genome of Apple chlorotic leaf spot virus. J Phytopathol 161:704–712
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Eriksson AE, Baase WA, Zhang XJ, Heinz DW, Blaber M, Baldwin EP, Matthews BW (1992) Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science 255:178–183
Gadiou S, Kundu JK, Paunovic S, Garcia-Diez P, Komorowska B, Gospodaryk A, Handa A, Massart S, Birisik N, Takur PD, Polischuk V (2010) Genetic diversity of flexiviruses infecting pome fruit trees. J Plant Pathol 92:685–691
Grimová L, Winkowska L, Zíka L, Ryšánek P (2016) Distribution of viruses in old commercial and abandoned orchards and wild apple trees. J Plant Pathol 98:549–554
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Guo W, Zheng W, Wang M, Li X, Ma Y, Dai H (2016) Genome sequences of three Apple chlorotic leaf spot virus isolates from hawthorns in China. PLoS ONE 11(8):e0161099
Huang X, Miller W (1991) A time-efficient, linear-spacer local similarity algorithm. Adv Appl Math 12:337–357
Jain R, Rivera MC, Lake JA (1999) Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci USA 96:3801–3806
Kinard GR, Scott SW, Barnett OB (1996) Detection of Apple chlorotic leaf spot and Apple stem grooving viruses using RT-PCR. Plant Dis 80:612–621
Krizbai L, Ember I, Németh M, Kölber M, Pasquini G, Faggioli F, Clark MF (2001) Characterization of Hungarian isolates of Apple chlorotic leaf spot virus. Acta Hortic 550:291–295
Li K, Shi HW, Jing CC, Sun XC, Zhou CY, Qing L (2015) Analysis of genome recombination and CP sequence diversity of ACLSV apple isolate from Shandong. Sci Agric Sin 48:2857–2867
Lister RM, Bancroft JB, Nadakavukaren MJ (1965) Some sap-transmissible viruses from apple. Phytopathology 55:859–870
Lu R, Folimonov A, Shintaku M, Li WX, Falk BW, Dawson WO, Ding SW (2004) Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc Natl Acad Sci U S A 101:15742–15747
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. https://doi.org/10.1093/ve/vev003
Mathioudakis MM, Maliogka VI, Katsiani AT, Katis NI (2010) Incidence and molecular variability of apple stem pitting and apple chlorotic leaf spot viruses in apple and pear orchards in Greece. J Plant Pathol 92:139–147
Mazeikiene I, Mozeryte D, Maliauskaite D, Siksnianiene JB (2012) Virologic status of the apple trees at the Institute of Horticulture. Sodininkystė ir daržininkystė. Sci Artic 3:13
Nagy PD (2008) Recombination in plant RNA viruses. In: Roossinck M (ed) Plant virus evolution. Springer, Berlin, pp 133–156
Nakahara KS, Yoshida K, Suzaki K, Yoshikawa N, Ito T (2011) Sensitive PCR based detection of Apple chlorotic leaf spot virus heterogeneous in apple trees. Jpn Agric Res Q 45:411–421
Nickel O, Silva FN, Fajardo TVM, Gorayeb ES (2018) Characterization and genetic variability of coat protein genes of Apple chlorotic leaf spot virus isolates from southern Brazil. Trop plant pathol 43(2):109–116
Niu F, Pan S, Wu Z, Jiang D, Li S (2012) Complete nucleotide sequences of the genomes of two isolates of apple chlorotic leaf spot virus from peach (Prunus persica) in China. Arch Virol 157:783–786
Pūpola N, Moročko-Bičevska I, Kāle A, Zeltiaš A (2011) Occurrence and diversity of pome fruit viruses in apple and pear orchards in Latvia. J Phytopathol 159:597–605
Rana T, Chandel V, Kumar Y, Ram R, Hallan V, Zaidi AA (2010) Molecular variability analyses of Apple chlorotic leaf spot virus capsid protein. J Biosci 35:605–615
Stankiene J, Mazeikiene I, Gelvonauskiene D, Sikšnianiene JB (2012) Virological status of stock planting material of apple and raspberry cultivars in Lithuania. Zemdirb Agric 99:93–98
Thomas CL, Leh V, Lederer C, Maule AJ (2003) Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306:33–41
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Verderevskaja TD, Marinescu VG (1985) Virus and mycoplasma diseases of fruit cultures and grapevine. Kishinev, Moldova, pp 117–127
Wang LP, Hong N, Mati S, Myrta A, Song YS, Michelutti R, Wang GP (2011) Pome fruit viruses at the Canadian Clonal Genebank and molecular characterization of Apple chlorotic leaf spot virus isolates. Sci Hortic 130:665–671
Waterworth H (1993) Processing foreign plant germplasm at the National Plant Germplasm Quarantine Center. Plant Dis 76:854–860
Yaegashi H, Isogai M, Tajima H, Sano T, Yoshikawa N (2007) Combinations of two amino acids (Ala40 and Phe75 or Ser40 and Tyr75) in the coat protein of Apple chlorotic leaf spot virus are crucial for infectivity. J Gen Virol 88:2611–2618
Yanase H (1974) Studies on apple latent viruses in Japan. Bull Fruit Trees Res Sta Jpn Ser C1:47–109
Funding
This work was carried out within the framework of the long-term research program ‘Horticulture: agro-biological basics and technologies’ implemented by Lithuanian Research Centre for Agriculture and Forestry.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Mazeikiene, I., Siksnianiene, J.B., Gelvonauskiene, D. et al. Prevalence and molecular variability of Apple chlorotic leaf spot virus capsid protein genes in Lithuania. J Plant Dis Prot 125, 389–396 (2018). https://doi.org/10.1007/s41348-018-0177-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41348-018-0177-1