Abstract
Inositol polyphosphates, in their water-soluble or lipid-bound forms, represent a large and multifaceted family of signalling molecules. Some inositol polyphosphates are well recognised as defining important signal transduction pathways, as in the case of the calcium release factor Ins(1,4,5)P3, generated by receptor activation-induced hydrolysis of the lipid PtdIns(4,5)P2 by phospholipase C. The birth of inositol polyphosphate research would not have occurred without the use of radioactive phosphate tracers that enabled the discovery of the “PI response”. Radioactive labels, mainly of phosphorus but also carbon and hydrogen (tritium), have been instrumental in the development of this research field and the establishment of the inositol polyphosphates as one of the most important networks of regulatory molecules present in eukaryotic cells. Advancements in microscopy and mass spectrometry and the development of colorimetric assays have facilitated inositol polyphosphate research, but have not eliminated the need for radioactive experimental approaches. In fact, such experiments have become easier with the cloning of the inositol polyphosphate kinases, enabling the systematic labelling of specific positions of the inositol ring with radioactive phosphate. This approach has been valuable for elucidating their metabolic pathways and identifying specific and novel functions for inositol polyphosphates. For example, the synthesis of radiolabelled inositol pyrophosphates has allowed the discovery of a new protein post-translational modification. Therefore, radioactive tracers have played and will continue to play an important role in dissecting the many complex aspects of inositol polyphosphate physiology. In this review we aim to highlight the historical importance of radioactivity in inositol polyphosphate research, as well as its modern usage.
Similar content being viewed by others
1 Introduction
Inositol polyphosphates comprise a vast and multifaceted family of cellular metabolites. The size of the family is explained by the ability to combinatorially substitute the six hydroxyls of the myo-inositol ring with phosphate moieties: mathematically, 64 such combinations are possible [1]. This number is in fact an underestimate, as diphosphate (or pyrophosphate) moieties also exist [2– 4]. Among these myriad inositol polyphosphates, without doubt the most famous is the calcium release factor Ins(1,4,5)P3, the prototypical second messenger. Hydrolysis of the lipid bond of PtdIns(4,5)P2 by phospholipase C (PLC) following receptor activation to release the water-soluble Ins(1,4,5)P3 and the lipid diacylglycerol (DAG) is a textbook example of signal transduction (Fig. 1) [5, 6]. It results in Ins(1,4,5)P3 binding to the InsP3 receptor and the consequent release of calcium from intracellular stores [7].
Schematic representation of the inositol cycle. Inositol acquired from the extracellular space is incorporated into lipids by the action of the phosphatidylinositol synthase (PI-synthase). The conversion of PtdIns (PI), first to PtdIns(4)P (PIP) and then to PtdIns(4,5)P2 (PIP2), generates the substrate for phospholipase C (PLC, boxed in red). Once receptor activation occurs, PLC generates two second messengers: the (plasma) membrane-resident diacylglycerol (DAG) and the calcium (Ca2+) release factor Ins(1,4,5)P3 (IP3). The latter is converted back to inositol via two dephosphorisation steps, closing the cycle. The inositol cycle is particularly active in stimulated mammalian cells. The Hokin “PI response” [8, 9] measures the [32P] taken up by the cell and its conversion to [32P] γATP, with the subsequent radioactive phosphorylation of DAG to phosphatidic acid (PA). PA is then reattached to inositol, creating radioactive PI. For graphical reasons, inositol is abbreviated here as “I” instead of “Ins”, and phosphatidylinositol as “PI” instead of “PtdIns”
This PLC activity was indirectly assayed during the first discovery of receptor-stimulated inositol polyphosphate metabolism. In the early 1950s, the Hokin husband and wife team were studying RNA metabolism using radioactive orthophosphate [32P] metabolic labelling, when they discovered an increase in cellular incorporation of radioactivity when pancreatic slices were stimulated with acetylcholine. Surprisingly, however, the large majority of radioactivity was not incorporated into nucleic acids but into the inositol-containing lipids called phosphoinositides (PI or PtdInsP, phosphatidylinositols); this acetylcholine-stimulated [32P] incorporation was therefore termed the “PI response” [8, 9]. It was not until three decades later that the PI response was determined to be part of PLC activation [6, 10] (Fig. 1). The [32P] taken up is initially incorporated into ATP and other nucleotides. Radioactive phosphorylation of DAG to phosphatidic acid (PA) allows its reattachment to inositol, creating PtdIns that is further phosphorylated, generating the radioactive PtdInsP/“PI” that the Hokins observed, and completing what is now known as the inositol cycle (Fig. 1).
This short historical background highlights the fundamental importance of radioactive phosphate labelling in the birth of the phosphoinositide and inositol polyphosphate signalling research fields. Equally, everyone is aware of the historical—and indeed, current—importance of radioactive labelling in nucleic acid research [11]. Without radioactivity-based methods, molecular biology would not have emerged and we would not be in the post-genomic era of biomedical research. It is not an overstatement to say that without radioactivity, the advances in biomedical science and consequent improvements in human health of the past century would not have been achieved. The ongoing importance of radioactive labelling to inositol polyphosphate research should also not be underestimated.
The general perception of “radioactivity” is one of fear, and understandably so, considering famous disasters such as those at Chernobyl or Fukushima [12]. However, using very low trace levels of radioactivity in a highly controlled research environment is safe: many protection and monitoring measures are available [13, 14]. Young researchers should therefore embrace radioactivity-based techniques, as they offer unique research opportunities, even in this twenty-first century, and especially in the inositol polyphosphate research field. Inositol polyphosphates, being unable to absorb UV/visible wavelength light, cannot be detected by absorbance or fluorescence methods. Thus the study of their metabolism and physiological functions has only been possible through radioactivity experiments. The current essay will focus primarily on the importance of radioactive phosphate labelling ([32P] and [33P]) in inositol polyphosphate biology. We will discuss the use of radioactive orthophosphate tracers to study the cellular metabolism of inositol polyphosphates, as well as the biochemical synthesis and purification of inositol polyphosphates radiolabelled at specific positions on the inositol ring. To provide a complete view of relevant radioactive methods, we will also concisely describe the use of radioactive hydrogen (tritium [3H]) and carbon (carbon-14 or [14C]) labelling techniques.
2 Phosphoinositides and Inositol Phosphates
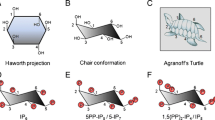
Before discussing radioactive labelling methods in inositol polyphosphate research, we must briefly introduce these molecules and the metabolic pathways connecting them. As it is not the main scope of this essay, discussion of inositol polyphosphate metabolism will be highly simplified; the interested person is encouraged to read the following more comprehensive reviews [15–18]. The carbon backbone of myo-inositol (hereafter simply called “inositol”) is by far the most common and biologically relevant of the naturally occurring stereoisomers. It is also the structural building block for the inositol polyphosphates. In its favoured chair conformation, inositol has five equatorial and one axial hydroxyl group (Fig. 2a) [19]. This axial hydroxyl is found at the carbon in position 2, using the D-numbering convention for cyclitols.
myo-Inositol structure and its radiolabelled derivatives. While nine stereoisomeric configurations of inositol are possible, the structure of myo-inositol is depicted in (a), referred to in the review simply as inositol, since it is by far the most common and biologically relevant form of inositol. The modern D-numbering system for inositols is counterclockwise as viewed from above and assigns the single axial hydroxyl group of myo-inositol to the carbon in position 2, while the other five hydroxyls are equatorial. myo-Inositol possesses an axis of symmetry through carbons 2 and 5 (dashed line), making positions 1,3 and 4,6 enantiomeric. The most common commercially available tritium-labelled inositol (b) possess the [3H] radiolabelled in position 1 and/or 2, while in [14C]inositol the radiolabel is uniformly distributed (c)
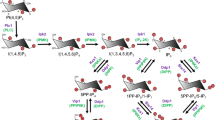
The simplest function of inositol is as an osmolyte, whose cellular concentration is regulated in response to hyperosmolarity. However, more interesting functions are achieved through phosphorylation to create inositol polyphosphates. These water-soluble molecules have a complicated biosynthetic pathway in yeast and, presumably, in mammalian cells: not just sequential phosphorylation or dephosphorylation, but synthesis that is intimately linked to the metabolism of the related PtdIns lipids (Figs. 1, 3) [20, 21]. Cells can synthesise inositol de novo from glucose-6-phosphate or acquire it from the extracellular environment, allowing it to enter the inositol cycle (Fig. 1). Inositol lipid synthesis starts with the activation of PA with CTP, becoming CDP-DAG that is subsequently attached to the 1-hydroxyl of the inositol ring, forming PtdIns. This can be phosphorylated to PtdIns(4)P and then to PtdIns(4,5)P2, the substrate for phospholipase C (PLC), in the calcium release signalling paradigm described above. It is important to remember that the universal production of Ins(1,4,5)P3 by PLC does not necessarily translate into calcium signalling, as many eukaryote clades including yeast and plants do not possess InsP3 receptors [22]. The Ins(1,4,5)P3 generated by PLC activity can be dephosphorylated back to inositol and reused for PtdIns synthesis. Conversely, Ins(1,4,5)P3 can act as precursor for a large, diverse family of higher phosphorylated inositol polyphosphates (Fig. 3). For example, Ins(1,4,5)P3 is a substrate of the inositol polyphosphate multikinase (IPMK; yeast Arg82) [23–26], which is able to phosphorylate both positions 3 and 6, creating Ins(1,3,4,5,6)P5. This is acted on by inositol pentakisphosphate 2-kinase (IP5-2K or IPPK; Ipk1 in yeast) [27, 28] to create the fully phosphorylated inositol hexakisphosphate (InsP6 or phytic acid). Another metabolic route can also lead to Ins(1,3,4,5,6)P5 synthesis. Phosphorylation of Ins(1,4,5)P3 by ITPKA,B,C, the IP3-3Ks, generates Ins(1,3,4,5)P4 [29, 30] that is dephosphorylated by 5-phosphatases such as SHIP1 into a different InsP3 isomer, Ins(1,3,4)P3. This isomer is a substrate for ITPK1, another multikinase, which adds a phosphate group at positions 5 and 6, again resulting in an InsP5 species with the remaining hydroxyl group at the 2 position [31–33] (Fig. 3). Furthermore ITPK1 can also phosphorylate position 1 of the inositol ring [34, 35].
Inositol polyphosphates synthetic pathway. The synthesis of higher phosphorylated inositol polyphosphates begins with the synthesis of Ins(1,4,5)P3. Saccharomyces cerevisiae uses only phospholipase C (PLC) hydrolysis of the lipid PI(4,5)P2 to synthesise Ins(1,4,5)P3 [28], whereas Dictyostelium discoideum utilises the cytosolic route, of which the enzymology is not fully elucidated (dashed line) [110]. Ins(1,4,5)P3 is metabolised by ITPKA, B, or C to synthesise Ins(1,3,4,5)P3, which is acted on by the 5-phosphatase (grey line) to generate the Ins(1,3,4)P3 converted by ITPK1 to Ins(1,3,4,5,6)P5. However, this isomer of InsP5 can also be directly generated by IPMK from Ins(1,4,5)P3. InsP5 is converted to InsP6 by the IP5-2Kinase IPPK. Phosphorylation of InsP6 by the IP6Ks generates the inositol pyrophosphate InsP7, specifically the depicted isomer 5PP-InsP5, which is further acted on by PPIP5K1,2 to IP8, specifically to 1,5(PP)2-IP4. The IP6K enzymes can also use Ins(1,3,4,5,6)P5 as a substrate, generating the inositol pyrophosphate PP-IP4. In this figure, for visual reasons, inositol is abbreviated as “I” instead of “Ins”, and phosphatidylinositol as “PI” instead “PtdIns”. Kinases catalysing each step are indicated in red (human) and blue (S. cerevisiae)
Together, Ins(1,3,4,5,6)P5 and InsP6 are the major forms of inositol polyphosphates present in mammalian cells. Despite their association with numerous cellular functions, and unlike Ins(1,4,5)P3, no dramatic or rapid changes in the cellular amount of either molecule are seen on receptor activation, although their relative level is modulated by neurotrophin signals [36]. However, they are not metabolically inert: Ins(1,3,4,5,6)P5 and InsP6 are substrates for the synthesis of a subfamily of inositol polyphosphates called inositol pyrophosphates [37–39]. These contain one or more high-energy phosphoanhydride (pyro) bonds as well as the phosphoesters. Two classes of enzyme synthesise inositol pyrophosphates in mammalian cells. The IP6Ks (InsP6 kinase) are able to pyrophosphorylate position 5 of the inositol ring, generating 5PP-InsP5 (diphosphoinositol pentakisphosphate, InsP7) from InsP6 or 5PP-InsP4 from Ins(1,3,4,5,6)P5 [25, 38–43]. Alternatively, PPIP5K (VIP1 in yeast) enzymes can pyrophosphorylate position 1, generating the 1PP-InsP5 isomer of InsP7 from InsP6 in vitro [44–47], and InsP8 ([PP]2-InsP4; bis-diphosphoinositol-tetrakisphosphate) from 5PP-InsP5 in vivo [48, 49].
The characterised inositol phosphate kinases (Table 1) offer the opportunity to label the inositol ring with radioactive phosphate simply by performing in vitro enzymatic reactions using recombinant enzyme with radioactive [32P]γATP. We will first introduce the radioactive isotopes available, before describing their use.
3 Radioactive Labels for Inositol
Inositol polyphosphates consist of four different atomic species: for example, the chemical formula of the fully phosphorylated inositol ring of InsP6 is C6H18O24P6. Of these elements, oxygen does not have relevant long-lived radioisotopes and therefore is not used for radiolabelling. Conversely, it is possible to generate useful unstable, and thus radioactive, isotopic species for carbon, hydrogen and phosphorus. We will briefly describe the biophysical characteristics of the most commonly used radioisotopes: tritium [3H], carbon [14C], and phosphorus [32P] and [33P] (Table 2). All are radioactive due to β decay.
Tritium contains two neutrons as well as the usual hydrogen proton. It is a weak β-emitter with a half-life of over 12 years. The emitted electron has very low energy, and can therefore travel only a few millimetres in air, and cannot penetrate the skin. This makes it particularly easy to work with, as special protective equipment is not required: gloves, goggles and lab coat are sufficient. On the minus side, the emitted radiation is too low to be detected by Geiger counter. Testing for tritium contamination before and after an experiment must instead be performed by swipe testing and liquid scintillation counting. Carbon-14 has a very long half-life, enabling its use in carbon dating in archaeology. The emitted electrons are still reasonably low-energy, although they can be detected using a Geiger counter, and can travel less than 0.3 mm into the skin. Therefore, shielding is also not necessary when working with [14C].
Phosphorus-32 is the most energetic radioisotope commonly used in biochemical laboratories. The electrons released are very high-energy and can travel over 6 m through air and 0.76 cm into human tissues. Consequently, extra precautions are required when working with [32P]: a 1 cm Plexiglas shield is required for the working area, and dosimeter monitoring for the experimenter. Contamination is easily detected with a Geiger counter. Experiments requiring [32P] can also be performed using the superior phosphorus-33 isotope. The emitted radiation from [33P] is of lower energy and is therefore less dangerous, although the same safety measures are required. The half-life of [33P] is also longer than that of [32P]: 25.4 days compared to 14.3 days. This can be extremely beneficial in maximising the value of any radiolabelled compounds synthesised. Only very rare experimental circumstances require the high energy of [32P] radiation, but this isotope is still widely used, since there is a huge price difference between the two radioactive isotopes. Phosphorus-33 is much more expensive.
4 Inositol Polyphosphate in Vivo Studies Using Radioactive Metabolic Labelling
As mentioned above, the main problem for inositol polyphosphate research is that there is no easy way to detect the inositol ring using spectrophotometry. A technique was developed in the 1980s that combined chromatography, post-column derivatisation and spectrophotometry to visualise the phosphate groups, and thus indirectly the inositol polyphosphate, to try to solve this problem [50]. This method is not sensitive enough to be routinely used with mammalian cells, although there are a few reports of its use [46]; it may be more appropriate for organisms with high levels of inositol polyphosphates such as the amoeba Dictyostelium discoideum [3, 51]. In general, for effective, sensitive and reliable methods for detecting and studying the metabolism and many functions of inositol polyphosphates, we must turn to radioactivity.
The standard procedures for investigating inositol polyphosphate metabolism in vivo require the use of radiolabelled inositol tracers [52, 53]. These are usually based on tritium labels, with carbon-14 rarely used, as it is more expensive (Fig. 2b, c). These [3H]inositol or [14C]inositol tracers (Fig. 2b, c) are added to the extracellular growth medium, where they are taken up by cells and enter the inositol cycle (Figs. 1, 3). Different inositol polyphosphates and phosphoinositides then begin to be radiolabelled. The labelling must be given sufficient time to reach isotopic equilibrium, where all the inositol polyphosphates species are in equilibrium with [3H]inositol or [14C]inositol. Given the presence of more than 30 inositol polyphosphate species in eukaryote cells [15, 17], and that, for example, seven sequential enzymatic reactions are required to generate InsP7, it is clear that allowing time to reach isotopic equilibrium is fundamental to generating reliable experimental data. For yeast, it is normally sufficient to label the cells overnight, which corresponds to 7–8 cell divisions. Unsurprisingly, mammalian cells must be labelled for much longer to reach equilibrium: 4 or 5 days may be enough, depending on cell type. Before starting labelling experiments in a new cell type, a pilot study must be performed to determine the time needed to reach metabolic equilibrium [36]. This is calculated by dividing the radioactivity accumulated in InsP6 by the radioactivity of the lipid phosphoinositide pool. This InsP6/phosphoinositide ratio increases over time; when it remains constant, the labelling has reached equilibrium. In the modern literature, too often the isotopic equilibrium is not properly calculated or even considered, casting doubt on the reliability of the data generated.
Once metabolic equilibrium has been reached, radiolabelled inositol polyphosphates can be acid-extracted and resolved by strong anion exchange high-performance liquid chromatography (saxHPLC). Two options exist for analysing the samples. Firstly, an in-line radioactivity detector can be used, greatly speeding up analysis and reducing handling at the cost of sensitivity. The alternative is to collect fractions for manual counting with a scintillation β-counter; while more labour-intensive, this method significantly increases the sensitivity. The inositol pyrophosphate species InsP7 and InsP8 can be easily detected in labelled yeast extracts using manual counting [52, 54], while only the more abundant precursor InsP6 has been identified using in-line detectors [28].
It should also be mentioned that dual isotopic labelling is possible. The difference in the energy of electrons emitted by [3H]inositol and [14C]inositol enables scintillation counters to distinguish these two inositol species. By labelling cells to isotopic equilibrium using [14C]inositol, and then briefly with [3H]inositol, it is possible to study the possible heterogeneity within pools of phosphoinositides or inositol polyphosphates [55]. This approach was employed successfully in the study of inositol polyphosphates generated after vasopressin or prostaglandin stimulation of vascular tissue. Their source was found to be rapidly labelled phosphoinositides, while the bulk of the highly phosphorylated inositol polyphosphates InsP5 and InsP6 were not created from the rapid phosphoinositide turnover, and were therefore deemed metabolically inert [56]. Dual labels can also have more technical uses. Chiefly, a second isotopic label is widely used to determine chromatographic saxHPLC peak identity: spiking a [3H]inositol labelled extract with a [14C]inositol polyphosphate standard enables conclusive identification of the nature of the eluted peaks [24].
The use of commercially available radiolabelled standards would of course be ideal. Unfortunately, many inositol polyphosphates, including InsP7, are not available commercially in their radiolabelled form or often even in unlabelled format. The next best option for peak identification is to use the well-characterised yeast inositol kinase mutant strains. Separation of radiolabelled wild-type yeast extracts by saxHPLC reveals a simple elution profile with one major peak of InsP6 and two smaller, more polar and therefore later-eluting peaks of InsP7 and InsP8. These two peaks are absent in extracts from kcs1Δ (IP6K mutant) yeast. The vip1Δ mutant instead accumulates InsP7 [48, 49]. Other mutants show increased peaks for inositol polyphosphates: Ins(1,4,5)P3 accumulates in arg82Δ multikinase mutants [23], while Ins(1,3,4,5,6)P5 accumulates and is converted by Kcs1 into PP-IP4 in an ipk1Δ mutant. The ipk1Δkcs1Δ double mutant has only the increased InsP5 peak [54]. Radiolabelled inositol polyphosphate standards can also be generated in vitro enzymatically, whether using radioactive inositol polyphosphate precursors or cold inositol polyphosphates with [32P]γATP (as described below). Laboratories with a serious interest in inositol polyphosphate analysis would do well to create standards with a long half-life: extracting and purifying specific inositol polyphosphates from [3H]inositol- or [14C]inositol-labelled cells gives defined standards that can be used over several years [24, 57].
While radioactive orthophosphate labelling has been instrumental in the early development [58–60] of the inositol phosphate research field, it must be admitted that [32P] or [33P] orthophosphate labelling currently has limited use for in vivo analysis of inositol polyphosphate metabolism. Many phosphorylated molecules in eukaryotic cells, primarily the abundant nucleotides, are co-purified during the acidic extraction normally employed to purify inositol polyphosphates. Therefore, any chromatogram is essentially undecipherable until the nucleotides have eluted off. The interfering nucleotides can be removed first by charcoal treatment, but this complicates the extraction procedure [61]. For this reason, only a handful of papers published after the 1990s have reported the use of phosphate labelling to study inositol polyphosphate metabolism in vivo. One such use was the original identification of inositol pyrophosphates in D. discoideum extracts [3]: the saxHPLC elution region for these high-polarity molecules in this organism is conveniently free from interference from contaminating phosphorylated molecules. This is not the case for extracts from cell types rich in the linear polymer of phosphates [62, 63] inorganic polyphosphate (polyP), such as yeast or trypanosomes, where polyP is particularly abundant [64–66]. In these organisms, orthophosphate labelling experiments will reveal the continuous presence throughout the chromatogram of radiolabelled polyP peaks that cover the InsP6, InsP7 and InsP8 signals (A. Saiardi, unpublished observation).
Orthophosphate labelling using [32P] or [33P] is more useful in studies of PtdIns lipids. While several types of potentially contaminating phospholipids exist, their number is not as great as the water-soluble (i.e., acid-extracted) phosphorylated molecules present in eukaryotic cytosol. Furthermore, nucleotides are eliminated by the organic solvent extraction procedure required to purify phosphoinositides. Thus it is possible to identify and study radiolabelled phosphoinositides directly, using thin-layer chromatography (TLC) or by resolution by saxHPLC after deacylation [67]. The highly energetic phosphate isotopes also allow the use of in-line radioactive detection methods to measure the saxHPLC-eluted deacylated lipids. It is important to note that orthophosphate [32P] or [33P] labelling is usually performed over a short period of time, from a few minutes to a few hours only. The phosphorus isotopes are toxic to the cell, and incubation over a few days induces cell stress or even death. Furthermore, orthophosphate labelling does not require reaching any isotopic equilibrium. Tritium or [14C] labels are components of the inositol ring itself (Fig. 1b, c), while phosphate labels are not: they are added and removed by the dynamic action of specific phosphatases and kinases, and thus isotopic equilibrium labelling is not necessary.
5 Biochemical Synthesis of 32/33Phosphate Radiolabelled Inositol Polyphosphates
Even before the cloning of inositol polyphosphate kinases, allowing the synthesis and purification of recombinant enzymes from bacteria, partially purified enzymatic activities or simple cell extracts were used to synthesise radiolabelled inositol polyphosphates [68, 69]. The synthesis of these molecules, labelled with radioactive phosphate at specific positions of the ring, has had numerous applications, including as standards, for monitoring enzymatic activity, or even the discovery of the new post-translational modification protein pyrophosphorylation. Most of the metabolic pathways and functions of lower phosphorylated inositol polyphosphates were elucidated before the cloning of the kinases responsible for their synthesis, often using elegant [32P] radiolabelled biochemical assays [70–73]. Studies of the functions of the higher phosphorylated inositol polyphosphates, such as InsP5, InsP6, and their inositol pyrophosphate derivatives have benefitted from this previous knowledge and by the cloning of their kinases. Using one of the six now known inositol polyphosphate kinases (Table 1) and [32/33P]γATP, it is possible to label almost any position of the inositol ring with radioactive phosphate. We will start by discussing the use of recombinant ITPKA, an IP3-3 kinase, to generate InsP4 labelled at position 3.
6 Synthesis of [32P]i Radiolabelled InsP4 and Its Use
The biosynthesis of [32P]InsP4 can be achieved by incubating ITPKA with Ins(1,4,5)P3 and [32P]γATP. This enzyme specifically phosphorylates position 3 of the ring, and thus generates 3[32P]Ins(1,3,4,5)P4 [74]. Several studies have used this method to investigate the metabolism of this isomer. Notably, its degradation by inositol polyphosphate 5-phosphatases (namely SHIP1/2) can be studied by following the formation of the radiolabelled and thus easily traceable 3[32P]Ins(1,3,4)P5 [75]. The anabolism of 3[32P]Ins(1,3,4,5)P4 and conversion to [32P]InsP6 has also been studied after incubation with nuclear extracts from D. discoideum [76].
SHIP2 is more famous for its ability to convert PtdIns(3,4,5)P3 to PtdIns(3,4)P2, thus regulating the signal from these two important lipids [77]. To study SHIP activity against lipids, radiolabelled [32P]PtdInsP3 substrate may be required. Previously, PI3Ks were employed to create this, but these are large proteins for which recombinant expression from bacteria is difficult. A current alternative is to use IPMK, which can act on not only the soluble InsP(1,4,5)P3 but also the lipid PtdIns(4,5)P2 [78], allowing straightforward synthesis of 3[32P]PtdIns(3,4,5)P3, specifically labelled in position 3 [79]. Human IPMK is easily produced from Escherichia coli [26, 78].
7 Preparing [32P]i Radiolabelled InsP5 and Its Use
Several inositol phosphate kinases are quite promiscuous. IPMK, as the name states, is a multikinase able to phosphorylate the inositol ring at positions 4 and 6, but also shows, at least in vitro, the ability to convert InsP5 to the inositol pyrophosphate PP-InsP4 [26, 80]. As noted above, it is also able to phosphorylate the lipid PtdIns(4,5)P2 to PtdIns(3,4,5)P3 [78, 81]. The IP6Ks can metabolise several isomers of InsP5 and InsP6 to inositol pyrophosphates [82, 83]. But perhaps the inositol phosphate kinase most catalytically flexible is ITPK1, which while primarily characterised as a 5- and 6-kinase (Fig. 3) [33], in certain species also has the ability to phosphorylate position 1 of the inositol ring [35]. This activity enables the synthesis of 1[32P]Ins(1,3,4,5,6)P5 [84] by incubating Ins(3,4,5,6)P4 with [32P]γATP in the presence of Entamoeba histolytica ITPK1 produced in E. coli [85] (Fig. 4). Using 1[32P]Ins(1,3,4,5,6)P5, an intriguing intersubstrate phosphate transfer activity was also discovered for ITPK1. In the presence of 1[32P]Ins(1,3,4,5,6)P5 and ADP, human ITPK1 transfers the radioactive phosphate, generating [32P]γATP and Ins(3,4,5,6)P4. The addition of Ins(1,3,4)P3 to this reaction augmented the rate of dephosphorylation of 1[32P]Ins(1,3,4,5,6)P5, as Ins(1,3,4)P3 now became the acceptor of the radioactive phosphate group, forming radiolabelled [32P]Ins(1,3,4,5/6)P4 phosphorylated at the 5 or 6 position, plus again Ins(3,4,5,6)P4 [84]. Thus the use of radioactive 1[32P]Ins(1,3,4,5,6)P5 enabled elucidation of how human ITPK1 regulates the synthesis of Ins(3,4,5,6)P4, a signalling molecule fundamental to controlling chloride channel conductance [31, 32]. It was later demonstrated, again using 1[32P]Ins(1,3,4,5,6)P5, that ITPK1 from the plant Solanum tuberosum possesses similar intersubstrate phosphotransferase activity [86].
Schematic synthesis of radiolabelled InsP5. The most abundant inositol pentakisphosphate isomer, Ins(1,3,4,5,6)P5, is the common end product of two multikinases, IPMK and ITPK1, using different starting inositol triphosphates (see Fig. 3). Therefore, using these two enzymes and different species of InsP3 and InsP4, it is possible to generate InsP5 labelled in different positions of the inositol ring. The top reaction illustrates the ability of Entamoeba histolytica ITPK1 (EhITPK1) to phosphorylate position 1 [34, 35], enabling specific synthesis of 1[32P]Ins(1,3,4,5,6)P5. Conversely, the bottom reaction uses the mammalian IPMK, a 3,6 kinase. Recombinant IPMK can thus be used to generate radiolabelled 3,6[32P]Ins(1,3,4,5,6)P5 [88]. Different atoms are colour-coded as follows: carbon black circle; oxygen green circle; phosphate yellow circle; radioactive phosphate red circle
A different [32P]Ins(1,3,4,5,6)P5 radiolabelled isomer was initially purified from metabolically labelled cells [72, 87]. It can now be made using IPMK with Ins(1,4,5)P3 and [32P]γATP, generating 3,6[32P]Ins(1,3,4,5,6)P5 (Fig. 4). Similarly, [3H]Ins(1,3,4,5,6)P3 is generated by incubating recombinant IPMK with the commercially available [3H]Ins(1,3,4,5)P4 and ATP. Both radiolabelled reagents 3,6[32P]Ins(1,3,4,5,6)P5 and [3H]Ins(1,3,4,5,6)P5 have been employed to great effect in studying the metabolic stability of InsP5 and its anti-tumour capability [88].
8 Preparing [32P]i Radiolabelled InsP6 and Its Use
The commercial availability of [3H]InsP6 has been intermittent over the years. This is unfortunate, as it was indispensable, for example, in the cloning of the IP6K enzymes from rat brain homogenate: the conversion of [3H]IP6 to [3H]InsP7 was followed using polyethylenimine cellulose thin-layer chromatography (PEI-TLC) [25, 68]. The custom synthesis of [32P]InsP6 was similarly essential to the identification of the other class of enzymes, PPIP5Ks, able to synthesise inositol pyrophosphates. High specific activity [32P]InsP6 can be generated enzymatically by incubating Ins(1,3,4,5,6)P5 and [32P]γATP with recombinant Arabidopsis thaliana IPK1 enzyme [89]. After HPLC purification, this enzymatic reaction generates [32P]IP6 specifically labelled in position 2, i.e., 2[32P]Ins(1,2,3,4,5,6)P6. The use of this compound spurred the identification, cloning and characterisation of the yeast PPIP5K (Vip1), since its conversion to [32P]InsP7 by kcs1Δ (IP6K deletion) yeast extracts revealed the presence of another inositol pyrophosphate synthase activity [47]. Radiolabelled [32P]InsP6 has also been used to follow IP6K activity during the yeast cell cycle [90] by simply resolving radioactive [32P]IP6 and [32P]IP7 by PEI-TLC. In addition to eliminating the need for a sophisticated HPLC apparatus, this experimental approach is quantitative, since the radioactivity present in the [32P]InsP6 and [32P]InsP7 TLC spots can be measured by scraping and counting them in a scintillation counter.
9 Preparing [32P]i Radiolabelled InsP7 and Its Use in Protein Pyrophosphorylation Reactions
The inositol pyrophosphates InsP7 and InsP8 have been linked to many cellular roles, but the mechanism is not clear. These are known to be dynamic molecules, unlike their “metabolically inert” precursor InsP6 [2]: in mammalian cells, up to 50% of the pool of InsP6 may be converted to InsP7 or InsP8 per hour [91]. One possible mode of action for InsP7 is protein pyrophosphorylation, in which the β-phosphate is donated to a pre-phosphorylated serine, becoming InsP6 and generating a pyrophosphoserine residue. The discovery of this post-translational modification followed the cloning of IP6K1 [25] and the subsequent ability to synthesise InsP7 radiolabelled at the β-position of the pyrophosphate moiety, 5[32P]βInsP7 (Fig. 5), using InsP6 and [32P]γATP [92]. Synthesis must be followed by a saxHPLC purification procedure to remove any leftover [32P]γATP. Radiolabelled 5[32P]βInsP7 fractions are then ready to use following desalting with a Sep-Pak QMA cartridge and concentration with a centrifugal evaporator [52]. Experiments demonstrated kinase-independent phosphorylation of multiple proteins in vitro [93]. In the absence of alternative detection methods, the use of the labelled 5[32P]β InsP7 is still reported in all publications of serine pyrophosphorylation [94–96].
Schematic synthesis of radiolabelled 5[32P]βInsP7. Two different InsP7 isomers can be easily and rapidly synthesised biochemically, using recombinant IP6K1 or the kinase domain of PPIP5K1 (or its yeast counterpart Vip1). Using radiolabelled [32P]γATP and InsP6 as substrate, IP6K1 generates 5[32P]βInsP7 (top), thus transferring the radioactive [32P] from ATP to the phosphorylated position 5 of InsP6. PPIP5K instead generates the isomer 1[32P]βInsP7 (bottom). Different atoms are colour-coded as follows: carbon black circle; oxygen green circle; phosphate yellow circle; radioactive phosphate red circle
Performing the in vitro pyrophosphorylation/transphosphorylation experiment, once 5[32P]βInsP7 is synthesised, is a straightforward process. However, it is important to remember the requirement for a pre-phosphorylated serine residue as substrate. Thus proteins of bacterial origin, such as recombinant mammalian proteins expressed in E. coli, require a priming event by a classical ATP kinase, usually casein kinase 2 (CK2) [93, 94]. The 5[32P]βInsP7 pyrophosphorylation is a non-enzymatic, temperature-dependent event that, while it does occur at physiological temperatures, is enhanced by incubating the sample at higher temperatures [97]. Once pyrophosphorylated, proteins can be resolved by polyacrylamide gel electrophoresis (PAGE), and their radioactivity incorporation can be easily identified by autoradiography. Several substrates of InsP7-mediated pyrophosphorylation have been identified so far. A common feature to all is stretches of serines embedded in regions rich in acidic residues; as magnesium is required for pyrophosphorylation, it is possible that the acidic residues are required to coordinate these ions. Pyrophosphorylated proteins are more acid-labile but also more resistant to phosphatases than proteins phosphorylated by ATP alone [93]. If this is true in vivo, it could have huge implications for cellular signalling: InsP7-mediated protein pyrophosphorylation could act in a dominant manner to allow continued signalling even during phosphatase activation.
It is important to note that inositol pyrophosphates appear to be interchangeable in their ability to pyrophosphorylate proteins. For example, 1[32P]βInsP7 synthesised using the Vip1 (PPIP5K) kinase domain and [32P]γATP (Fig. 5) is able to pyrophosphorylate proteins, as is 1,5[32P]InsP8 synthesised by the double action of IP6K1 and Vip1. In fact, the incubation of S. cerevisiae extracts with 5[32P]βInsP7, 1[32P]βInsP7 or 1,5[32P]βInsP8 revealed an identical pattern of pyrophosphorylated proteins [93]. This indicates that if pyrophosphorylation is the mechanism of action for inositol pyrophosphates, they can substitute for each other in performing their biological role. However, allosteric regulation of proteins has also been proposed as a mechanism [98, 99], and in this circumstance, different isomeric species of inositol pyrophosphates could be recognised by specific effector proteins.
The synthesis described here for 5[32P]βInsP7 is a straightforward procedure, as is using it to identify pyrophosphorylated proteins. However, processing millicurie amounts of radioactivity to generate a reagent with a two week half-life requires a degree of dedication. Therefore, protein pyrophosphorylation, while considered an exciting post-translational modification, has received little attention, as only a handful of laboratories have invested in the synthesis of 5[32P]βInsP7. The recent development of organic synthesised experimental tools to study protein pyrophosphorylation [100, 101] will likely lead to the further and very welcome development of non-radioactive detection methods, and to the demonstration of the existence of this modification in vivo. These developments will certainly increase interest in protein pyrophosphorylation, but it must be remembered that it was the innovative use of radioactive labelling methods that permitted the discovery of this modification in the first place. This is an excellent demonstration that original radioactive assays should not be avoided.
10 Perspective
In today’s “omics” era, radioactive labelling appears anachronistic. Current proteomic, metabolomic or genomic studies give us plentiful information in an ever-growing number of databases. Ultimately, however, these huge amounts of data must not be just statistically annotated, but must drive biochemistry experiments, which as noted before, provide the only experimental basis for the understanding of biological mechanisms [102, 103]. In this context, radioactive molecular tracers have played and will continue to play a contributory role in our quest to understand the molecular mechanisms of life.
This review has highlighted the fundamental importance of radioactive labelling in the birth and the development of inositol polyphosphate research. Although recent technological development efforts such as PAGE analysis [82] and TiO2 extraction of inositol polyphosphates [104] are facilitating the study of the metabolism and functions of these molecules without the need for radioactive precursors or metabolic labelling, we definitely foresee further need for phosphate-32/33-, carbon-14- or tritium-based experiments to fully appreciate the importance of these molecules in cell biology.
References
York JD (2006) Regulation of nuclear processes by inositol polyphosphates. Biochim Biophys Acta 1761(5–6):552–559
Menniti FS et al (1993) Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J Biol Chem 268(6):3850–3856
Stephens L et al (1993) The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s). J Biol Chem 268(6):4009–4015
Bennett M et al (2006) Inositol pyrophosphates: metabolism and signaling. Cell Mol Life Sci 63(5):552–564
Irvine RF (2003) 20 years of Ins(1,4,5)P3, and 40 years before. Nat Rev Mol Cell Biol 4(7):586–590
Michell RH (1975) Inositol phospholipids and cell surface receptor function. Biochim Biophys Acta 415(1):81–147
Berridge MJ, Lipp P, Bootman MD (2000) The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol 1(1):11–21
Hokin LE, Hokin MR (1955) Effects of acetylcholine on phosphate turnover in phospholipides of brain cortex in vitro. Biochim Biophys Acta 16(2):229–237
Hokin MR, Hokin LE (1953) Enzyme secretion and the incorporation of P32 into phospholipides of pancreas slices. J Biol Chem 203(2):967–977
Streb H et al (1983) Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate. Nature 306(5938):67–69
Hershey AD, Chase M (1952) Independent functions of viral protein and nucleic acid in growth of bacteriophage. J Gen Physiol 36(1):39–56
Ivanov VK (2012) Lessons from Chernobyl and prognosis for Fukushima: radiological consequences. J Radiol Prot 32(1):N55–N58
Engelbrecht R, Schwaiger M (2008) State of the art of standard methods used for environmental radioactivity monitoring. Appl Radiat Isot 66(11):1604–1610
Meisenhelder J, Semba K (2001) Safe use of radioisotopes. Curr Protoc Cell Biol Appendix 1: p. Appendix 1D
Irvine RF, Schell MJ (2001) Back in the water: the return of the inositol phosphates. Nat Rev Mol Cell Biol 2(5):327–338
Resnick AC, Saiardi A (2008) Inositol Polyphosphates. In: Begley TP (ed) Wiley Encyclopedia of Chemical Biology. Wiley, Hoboken
Shears SB et al (2012) Defining signal transduction by inositol phosphates. Subcell Biochem 59:389–412
York JD et al (2001) An expanded view of inositol signaling. Adv Enzyme Regul 41:57–71
Irvine RF (2005) Inositide evolution-towards turtle domination? J Physiol 566(Pt 2):295–300
Balla T (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev 93(3):1019–1137
Di Paolo G, De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443(7112):651–657
Livermore TM et al (2016) Phosphate, inositol and polyphosphates. Biochem Soc Trans 44(1):253–259
Odom AR et al (2000) A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287(5460):2026–2029
Saiardi A et al (2000) Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett 468(1):28–32
Saiardi A et al (1999) Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol 9(22):1323–1326
Saiardi A et al (2001) Mammalian inositol polyphosphate multikinase synthesizes inositol 1,4,5-trisphosphate and an inositol pyrophosphate. Proc Natl Acad Sci U S A 98(5):2306–2311
Verbsky JW et al (2002) The synthesis of inositol hexakisphosphate. Characterization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. J Biol Chem 277(35):31857–31862
York JD et al (1999) A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285(5424):96–100
Takazawa K et al (1991) Molecular cloning and expression of a human brain inositol 1,4,5-trisphosphate 3-kinase. Biochem Biophys Res Commun 174(2):529–535
Togashi S et al (1997) Structural identification of the myo-inositol 1,4,5-trisphosphate-binding domain in rat brain inositol 1,4,5-trisphosphate 3-kinase. Biochem J 326(Pt 1):221–225
Mitchell J et al (2008) An expanded biological repertoire for Ins(3,4,5,6)P4 through its modulation of ClC-3 function. Curr Biol 18(20):1600–1605
Saiardi A, Cockcroft S (2008) Human ITPK1: a reversible inositol phosphate kinase/phosphatase that links receptor-dependent phospholipase C to Ca2+-activated chloride channels. Sci Signal 1(4):pe5
Wilson MP, Majerus PW (1996) Isolation of inositol 1,3,4-trisphosphate 5/6-kinase, cDNA cloning and expression of the recombinant enzyme. J Biol Chem 271(20):11904–11910
Liu C et al (2001) Synthesis and biological activity of D- and L-chiro-inositol 2,3,4,5-tetrakisphosphate: design of a novel and potent inhibitor of Ins(3,4,5,6)P4 1-kinase/Ins(1,3,4)P3 5/6-kinase. J Med Chem 44(18):2984–2989
Yang X, Shears SB (2000) Multitasking in signal transduction by a promiscuous human Ins(3,4,5,6)P(4) 1-kinase/Ins(1,3,4)P(3) 5/6-kinase. Biochem J 351(Pt 3):551–555
Loss O et al (2013) Modulation of inositol polyphosphate levels regulates neuronal differentiation. Mol Biol Cell 24(18):2981–2989
Shears SB (2009) Diphosphoinositol polyphosphates: metabolic messengers? Mol Pharmacol 76(2):236–252
Wilson MS, Livermore TM, Saiardi A (2013) Inositol pyrophosphates: between signalling and metabolism. Biochem J 452(3):369–379
Wundenberg T, Mayr GW (2012) Synthesis and biological actions of diphosphoinositol phosphates (inositol pyrophosphates), regulators of cell homeostasis. Biol Chem 393(9):979–998
Barker CJ et al (2009) Inositol pyrophosphates: structure, enzymology and function. Cell Mol Life Sci 66(24):3851–3871
Draskovic P et al (2008) Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem Biol 15(3):274–286
Saiardi A et al (2001) Identification and characterization of a novel inositol hexakisphosphate kinase. J Biol Chem 276(42):39179–39185
Shears SB (2015) Inositol pyrophosphates: why so many phosphates? Adv Biol Regul 57:203–216
Choi JH et al (2007) Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J Biol Chem 282(42):30763–30775
Fridy PC et al (2007) Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J Biol Chem 282(42):30754–30762
Lin H et al (2009) Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J Biol Chem 284(3):1863–1872
Mulugu S et al (2007) A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316(5821):106–109
Laha D et al (2015) VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and jasmonate-dependent defenses in Arabidopsis. Plant Cell 27(4):1082–1097
Onnebo SM, Saiardi A (2009) Inositol pyrophosphates modulate hydrogen peroxide signaling. Biochem J 423(1):109–118. doi:10.1042/BJ20090241
Mayr GW (1988) A novel metal-dye detection system permits picomolar-range h.p.l.c. analysis of inositol polyphosphates from non-radioactively labelled cell or tissue specimens. Biochem J 254(2):585–591
Albert C et al (1997) Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem J 327(Pt 2):553–560
Azevedo C, Saiardi A (2006) Extraction and analysis of soluble inositol polyphosphates from yeast. Nat Protoc 1(5):2416–2422
Stevenson-Paulik J et al (2006) Inositol phosphate metabolomics: merging genetic perturbation with modernized radiolabeling methods. Methods 39(2):112–121
Saiardi A et al (2002) Inositol pyrophosphates regulate endocytic trafficking. Proc Natl Acad Sci U S A 99(22):14206–14211
Michell RH et al (1988) Inositol lipids: receptor-stimulated hydrolysis and cellular lipid pools. Philos Trans R Soc Lond B Biol Sci 320(1199):239–246
Maccallum SH et al (1989) The use of cells doubly labelled with [14C]inositol and [3H]inositol to search for a hormone-sensitive inositol lipid pool with atypically rapid metabolic turnover. J Endocrinol 122(1):379–389
Hughes PJ et al (1989) The regulation of the phosphorylation of inositol 1,3,4-trisphosphate in cell-free preparations and its relevance to the formation of inositol 1,3,4,6-tetrakisphosphate in agonist-stimulated rat parotid acinar cells. J Biol Chem 264(33):19871–19878
Downes CP, Hawkins PT, Irvine RF (1986) Inositol 1,3,4,5-tetrakisphosphate and not phosphatidylinositol 3,4-bisphosphate is the probable precursor of inositol 1,3,4-trisphosphate in agonist-stimulated parotid gland. Biochem J 238(2):501–506
King CE et al (1987) Multiple metabolic pools of phosphoinositides and phosphatidate in human erythrocytes incubated in a medium that permits rapid transmembrane exchange of phosphate. Biochem J 244(1):209–217
Palmer S et al (1986) The labelling of polyphosphoinositides with [32P]Pi and the accumulation of inositol phosphates in vasopressin-stimulated hepatocytes. Biochem J 238(2):491–499
Sulpice JC et al (1989) The separation of [32P]inositol phosphates by ion-pair chromatography: optimization of the method and biological applications. Anal Biochem 179(1):90–97
Azevedo C, Saiardi A (2014) Functions of inorganic polyphosphates in eukaryotic cells: a coat of many colours. Biochem Soc Trans 42(1):98–102
Docampo R, Ulrich P, Moreno SN (2010) Evolution of acidocalcisomes and their role in polyphosphate storage and osmoregulation in eukaryotic microbes. Philos Trans R Soc Lond B Biol Sci 365(1541):775–784
Gerasimaite R et al (2014) Coupled synthesis and translocation restrains polyphosphate to acidocalcisome-like vacuoles and prevents its toxicity. J Cell Sci 127(Pt 23):5093–5104
Lander N, Ulrich PN, Docampo R (2013) Trypanosoma brucei vacuolar transporter chaperone 4 (TbVtc4) is an acidocalcisome polyphosphate kinase required for in vivo infection. J Biol Chem 288(47):34205–34216
Lonetti A et al (2011) Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J Biol Chem 286(37):31966–31974
Jones DR et al (2013) Measurement of phosphoinositides in the zebrafish Danio rerio. Nat Protoc 8(6):1058–1072
Voglmaier SM et al (1996) Purified inositol hexakisphosphate kinase is an ATP synthase: diphosphoinositol pentakisphosphate as a high-energy phosphate donor. Proc Natl Acad Sci U S A 93(9):4305–4310
Ye W et al (1995) Inhibition of clathrin assembly by high affinity binding of specific inositol polyphosphates to the synapse-specific clathrin assembly protein AP-3. J Biol Chem 270(4):1564–1568
Downes CP, Michell RH (1981) The polyphosphoinositide phosphodiesterase of erythrocyte membranes. Biochem J 198(1):133–140
Shears SB et al (1987) Dephosphorylation of myo-inositol 1,4,5-trisphosphate and myo-inositol 1,3,4-triphosphate. Biochem J 242(2):393–402
Stephens LR, Downes CP (1990) Product-precursor relationships amongst inositol polyphosphates. Incorporation of [32P]Pi into myo-inositol 1,3,4,6-tetrakisphosphate, myo-inositol 1,3,4,5-tetrakisphosphate, myo-inositol 3,4,5,6-tetrakisphosphate and myo-inositol 1,3,4,5,6-pentakisphosphate in intact avian erythrocytes. Biochem J 265(2):435–452
Stephens LR et al (1988) L-myo-inositol 1,4,5,6-tetrakisphosphate (3-hydroxy)kinase. Biochem J 249(1):283–292
Van Dijken P et al (1994) Phosphorylation of inositol 1,4,5-trisphosphate analogues by 3-kinase and dephosphorylation of inositol 1,3,4,5-tetrakisphosphate analogues by 5-phosphatase. Eur J Biochem 226(2):561–566
Pesesse X et al (1998) The SH2 domain containing inositol 5-phosphatase SHIP2 displays phosphatidylinositol 3,4,5-trisphosphate and inositol 1,3,4,5-tetrakisphosphate 5-phosphatase activity. FEBS Lett 437(3):301–303
Van der Kaay J, Wesseling J, Van Haastert PJ (1995) Nucleus-associated phosphorylation of Ins(1,4,5)P3 to InsP6 in Dictyostelium. Biochem J 312(Pt 3):911–917
Erneux C et al (2011) SHIP2 multiple functions: a balance between a negative control of PtdIns(3,4,5)P(3) level, a positive control of PtdIns(3,4)P(2) production, and intrinsic docking properties. J Cell Biochem 112(9):2203–2209
Resnick AC et al (2005) Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc Natl Acad Sci U S A 102(36):12783–12788
Vandeput F et al (2006) The influence of anionic lipids on SHIP2 phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase activity. Cell Signal 18(12):2193–2199
Zhang T, Caffrey JJ, Shears SB (2001) The transcriptional regulator, Arg82, is a hybrid kinase with both monophosphoinositol and diphosphoinositol polyphosphate synthase activity. FEBS Lett 494(3):208–212
Maag D et al (2011) Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc Natl Acad Sci U S A 108(4):1391–1396
Losito O et al (2009) Inositol pyrophosphates and their unique metabolic complexity: analysis by gel electrophoresis. PLoS One 4(5):e5580
Saiardi A et al (2000) The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J Biol Chem 275(32):24686–24692
Chamberlain PP et al (2007) Integration of inositol phosphate signaling pathways via human ITPK1. J Biol Chem 282(38):28117–28125
Miller GJ et al (2005) Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase. Mol Cell 18(2):201–212
Caddick SE et al (2008) A Solanum tuberosum inositol phosphate kinase (StITPK1) displaying inositol phosphate-inositol phosphate and inositol phosphate-ADP phosphotransferase activities. FEBS Lett 582(12):1731–1737
Caffrey JJ et al (2001) Expanding coincident signaling by PTEN through its inositol 1,3,4,5,6-pentakisphosphate 3-phosphatase activity. FEBS Lett 499(1–2):6–10
Maffucci T et al (2005) Inhibition of the phosphatidylinositol 3-kinase/akt pathway by inositol pentakisphosphate results in antiangiogenic and antitumor effects. Cancer Res 65(18):8339–8349
Stevenson-Paulik J et al (2005) Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc Natl Acad Sci U S A 102(35):12612–12617
Banfic H et al (2013) Inositol pyrophosphates modulate S phase progression after pheromone-induced arrest in Saccharomyces cerevisiae. J Biol Chem 288(3):1717–1725
Glennon MC, Shears SB (1993) Turnover of inositol pentakisphosphates, inositol hexakisphosphate and diphosphoinositol polyphosphates in primary cultured hepatocytes. Biochem J 293(Pt 2):583–590
Saiardi A et al (2004) Phosphorylation of proteins by inositol pyrophosphates. Science 306(5704):2101–2105
Bhandari R et al (2007) Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc Natl Acad Sci U S A 104(39):15305–15310
Azevedo C et al (2009) Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc Natl Acad Sci U S A 106(50):21161–21166
Chanduri M et al (2016) Inositol hexakisphosphate kinase 1 (IP6K1) activity is required for cytoplasmic dynein-driven transport. Biochem J 473(19):3031–3047
Thota SG et al (2015) Inositol pyrophosphates regulate RNA polymerase I-mediated rRNA transcription in Saccharomyces cerevisiae. Biochem J 466(1):105–114
Werner Jr JK, Speed T, Bhandari R (2010) Protein pyrophosphorylation by diphosphoinositol pentakisphosphate (InsP7). Methods Mol Biol 645:87–102
Burton A, Hu X, Saiardi A (2009) Are inositol pyrophosphates signalling molecules? J Cell Physiol 220(1):8–15
Shears SB et al (2011) Diphosphoinositol polyphosphates: what are the mechanisms? Adv Enzyme Regul 51(1):13–25
Brown NW, Marmelstein AM, Fiedler D (2016) Chemical tools for interrogating inositol pyrophosphate structure and function. Chem Soc Rev 45(22):6311–6326
Wu M et al (2014) Elucidating diphosphoinositol polyphosphate function with nonhydrolyzable analogues. Angew Chem Int Ed Engl 53(28):7192–7197
Brenner S (2000) Biochemistry strikes back. Trends Biochem Sci 25(12):584
Kornberg A (2004) Biochemistry matters. Nat Struct Mol Biol 11(6):493
Wilson MS et al (2015) A novel method for the purification of inositol phosphates from biological samples reveals that no phytate is present in human plasma or urine. Open Biol 5(3):150014
Communi D, Vanweyenberg V, Erneux C (1995) Molecular study and regulation of D-myo-inositol 1,4,5-trisphosphate 3-kinase. Cell Signal 7(7):643–650
Dewaste V et al (2000) Cloning and expression of a cDNA encoding human inositol 1,4,5-trisphosphate 3-kinase C. Biochem J 352(Pt 2):343–351
Takazawa K et al (1990) Cloning and expression in E. coli of a rat brain cDNA encoding a Ca2+/calmodulin-sensitive inositol 1,4,5-trisphosphate 3-kinase. Biochem J 272(1):107–112
Wilson MP et al (2001) Inositol 1,3,4-trisphosphate 5/6-kinase is a protein kinase that phosphorylates the transcription factors c-Jun and ATF-2. J Biol Chem 276(44):40998–41004
Wang H et al (2012) Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat Chem Biol 8(1):111–116
Stephens LR, Irvine RF (1990) Stepwise phosphorylation of myo-inositol leading to myo-inositol hexakisphosphate in Dictyostelium. Nature 346(6284):580–583
Acknowledgements
We wish to thank the members of the Saiardi laboratory for reading of the manuscript. We have tried to review all the relevant literature, but we may have missed some specific work; therefore, we would like to apologise to colleagues whose research was not properly recognised. This work is supported by the Medical Research Council (MRC) core support to the MRC/UCL Laboratory for Molecular Cell Biology University Unit (MC_U122680443).
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection “Phosphate Labelling in Chemical Biology”; edited by Henning Jessen.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wilson, M.S.C., Saiardi, A. Importance of Radioactive Labelling to Elucidate Inositol Polyphosphate Signalling. Top Curr Chem (Z) 375, 14 (2017). https://doi.org/10.1007/s41061-016-0099-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41061-016-0099-y