Abstract
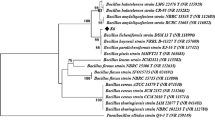
Bacterial cellulose (BC) is an extremely pure and highly valuable biomaterial. However, its production cost poses a challenge for large-scale manufacturing. This study explores a cost-effective approach by co-cultivating lactic acid bacteria with cellulose-synthesizing bacteria. Four BC-producing isolates from spoiled fruits and four lactic acid bacteria from fermented buttermilk were isolated and characterized. Growth studies demonstrated successful lactic acid bacteria cultivation in HS media. Co-cultivation of cellulose-synthesizing bacteria and lactic acid bacteria showed enhanced BC production, with a twofold increase in dry weight (0.35 g/150 ml) compared to the monoculture of cellulose-synthesizing bacteria (0.15 g/150 ml). Scanning electron microscopy revealed improved BC microfibril quality in co-culture. Reversed-phase HPLC confirmed higher lactic acid concentrations in co-culture. 16S rRNA sequence analysis revealed that lactic acid bacteria had a 100% match with Lactococcus lactis. These findings highlight the potential of co-cultivation for cost-effective BC production and lactic acid yield, offering a sustainable approach to biomaterial production.










Similar content being viewed by others
Abbreviations
- BC:
-
Bacterial cellulose
- MRS:
-
DeMan, Rogosa and Sharpe
- HS:
-
Hestrin and Schramm
- LAB:
-
Lactic acid bacteria
- CSB:
-
Cellulose-synthesizing bacteria
References
Barman S, Ghosh R, Mandal NC (2018) Production optimization of broad spectrum bacteriocin of three strains of Lactococcus lactis isolated from homemade buttermilk. Ann Agrar Sci 16(3):286–296
Barthalomew JW, Mittewer T (1950) A simplified bacterial stain. Stain Technol 25:153
De Man JC, Rogosa D, Sharpe ME (1960) A medium for the cultivation of Lactobacilli. J Appl Bacteriol 23(1):130–135
Hestrin S, Schramm MJBJ (1954) Synthesis of cellulose by Acetobacter xylinum. 2. preparation of freeze-dried cells capable of polymerizing glucose to cellulose. Biochem J 58(2):345–352
Indriyati, Dara F, Primadona I, Srikandace Y, Karina M (2021) Development of bacterial cellulose/chitosan films: structural, physicochemical and antimicrobial properties. J Polym Res. https://doi.org/10.1007/s10965-020-02328-6
Jang WD, Hwang JH, Kim HU, Ryu JY, Lee SY (2017) Bacterial cellulose as an example product for sustainable production and consumption. Microb Biotechnol 10(5):1181
Jayashree S, Pushpanathan M, Rajendhran J, Gunasekaran P (2013) Microbial diversity and phylogeny analysis of buttermilk, a fermented milk product, employing 16S rRNA-based pyrosequencing. Food Biotechnol 27(3):213–221
Lee KY, Buldum G, Mantalaris A, Bismarck A (2014) More than meets the eye in bacterial cellulose: biosynthesis, bioprocessing, and applications in advanced fiber composites. Macromol Biosci 14(1):10–32
Lotfy VF, Basta AH, Abdel-Monem MO, Abdel-Hamed GZ (2021) Utilization of bacteria in rotten guava for production of bacterial cellulose from isolated and protein waste. Carbohydr Polym Technol Appl 2:100076
Lu H, Jia Q, Chen L, Zhang L (2016) Effect of organic acids on bacterial cellulose produced by Acetobacter xylinum. J Microbiol Biotechnol 5(2):1–6
Maslienko LV, Voronkova AK, Datsenko LA, Efimtseva EA, Punogina DA, Gaydukova SA, Kazakova VV, Kovalyova SR (2020) The primary screening of fungal strains antagonists from a collection of the biological methods laboratory in VNIIMK to a phoma rot on sunflower. Part II Oil Crops 3(183):107–113
Mateo JJ, Maicas S (2015) Valorization of winery and oil mill wastes by microbial technologies. Food Res Int 73:13–25
Pasaribu KM, Ilyas S, Tamrin T, Radecka I, Swingler S, Gupta A, Stamboulis AG, Gea S (2023) Bioactive bacterial cellulose wound dressings for burns with collagen in-situ and chitosan ex-situ impregnation. Int J Biol Macromol 230:123118
Phomrak S, Phisalaphong M (2020) Lactic acid modified natural rubber–bacterial cellulose composites. Appl Sci 10(10):3583
Płoska J, Garbowska M, Klempová S, Stasiak-Różańska L (2023) Obtaining bacterial cellulose through selected strains of acetic acid bacteria in classical and waste media. Appl Sci 13(11):6429
Rahman SSA, Vaishnavi T, Vidyasri GS, Sathya K, Priyanka P, Venkatachalam P, Karuppiah S (2021) Production of bacterial cellulose using Gluconacetobacter kombuchae immobilized on Luffa aegyptiaca support. Sci Rep 11(1):2912
Saleh AK, El-Gendi H, Soliman NA, El-Zawawy WK, Abdel-Fattah YR (2022) Bioprocess development for bacterial cellulose biosynthesis by novel Lactiplantibacillus plantarum isolate along with characterization and antimicrobial assessment of fabricated membrane. Sci Rep 12(1):2181
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38(7):3022–3027
Thongwai N, Futui W, Ladpala N, Sirichai B, Weechan A, Kanklai J, Rungsirivanich P (2022) Characterization of bacterial cellulose produced by Komagataeibacter maltaceti P285 isolated from contaminated honey wine. Microorganisms 10(3):528
Toda K, Asakura T, Fukaya M, Entani E, Kawamura Y (1997) Cellulose production by acetic acid-resistant Acetobacter xylinum. J Ferment Bioeng 84(3):228–231
Volova TG, Prudnikova SV, Sukovatyi AG, Shishatskaya EI (2018) Production and properties of bacterial cellulose by the strain Komagataeibacter xylinus B-12068. Appl Microbiol Biotechnol 102(17):7417–7428
Wang ZG (2008) Effect of organic acids on bacterial cellulose production in defined medium by Acetobacter xylinum. Food Sci 29:295–297
Zeng A, Yang R, Tong Y, Zhao W (2023) Functional bacterial cellulose nanofibrils with silver nanoparticles and its antibacterial application. Int J Biol Macromol 235:123739
Acknowledgements
The authors would like to express their gratitude to the PG Department of Biotechnology at Alva’s College for providing all essential materials and equipment for carrying out this work.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Akki, A.J., Hiremath, L.D. & Badkillaya, R.R. Harnessing Symbiotic Association of Lactic Acid Bacteria and Cellulose-Synthesizing Bacteria for Enhanced Biological Activity. Iran J Sci 48, 311–320 (2024). https://doi.org/10.1007/s40995-023-01567-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40995-023-01567-8