Abstract
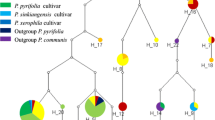
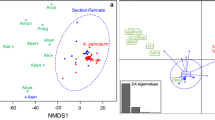
Date palm (Phoenix dactylifera L.) is a member of the genus Phoenix in Arecaceae family. It is the cultivated species in this genus with great economical and social importance in our country. We used three chloroplast regions of rbcL, matK, and trnH‐psbA with the following aims: 1. To illustrate the cultivars phylogeny, 2. to identify nucleotide variations and identify the molecular events leading to these variations, and finally, 3. to provide DNA barcode for the studied cultivars. ILD (incongruence length difference test) heterogeneity test revealed that the genes used are not homogenous and produce different evolutionary signals, and the phylogenetic trees constructed by each of these genes differ in topology. A combined data were used to produce phylogenetic tree by SVDQuartets method, to consider evolutionary signals of all three genes together. The SVDQuartets tree and Bayesian-based phylogenetic tree obtained from BEAST were utilized to construct a condensed tree and produce divergence time by RASP program. Date palm cultivars were separated in two distinct major clades corresponding to their geographical distribution, Khuzestan province and Sistan and Baluchestan province. The present study produced some new information with regard to potential DNA barcodes for commercial date palms cultivation and threw light on the date of cultivar divergence in this area.




Similar content being viewed by others
Availability of Data and Materials
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Abbreviations
- rbcl :
-
Ribulose bisphosphate carboxylase large chain
- matk :
-
Maturase K
- SSR:
-
Simple sequence repeats
- ISSR:
-
Inter simple sequence repeats
- SCoT:
-
Start codon targeted
- RAPD:
-
Randomly amplified polymorphic DNA
- ITS:
-
Internal transcribed spacer
- ML:
-
Maximum likelihood tree
- TCS:
-
Transitive consistency score
- DCA:
-
Dentrented correspondence analysis
- GTR + G:
-
General time reversible + gamma distribution
- AMOVA:
-
Analysis of molecular variance
References
Al-Qurainy F, Khan S, Al-Hemaid FM, AliMA TM, Ashraf M (2011) Assessing molecular signature for some potential date (Phoenix dactylifera L.) cultivars from Saudi Arabia, based on chloroplast DNA sequences rpoB and psbA-trnH. Int J Mol Sci 12:6871–6880. https://doi.org/10.3390/ijms12106871
Bahraminejad A, Mohammadi-Nejad G (2015) Genetic diversity analysis of date palm (Phoenix dactylifera L.) genotypes using RAPD markers. Ann Res Rev Biol. https://doi.org/10.9734/ARRB/2015/11683
Chao CT, Krueger RR (2007) The date palm (Phoenix dactylifera L.): overview of biology uses and cultivation. Hort Sci 42:1077–1082. https://doi.org/10.21273/HORTSCI.42.5.1077
Chou J, Gupta A, Yaduvanshi S, Davidson R, Nute M, Mirarab S, Warnow T (2015) A comparative study of SVDquartets and other coalescent-based species tree estimation methods. BMC Genom 16(10):S2
Flowers JM, HazzouriK M, Gros-Balthazard M, Mo Z, Koutroumpa K, Perrakis A, Ferrand S, Khierallah HSM, Fuller DQ, Aberlenc F, Fournaraki C, Purugganan MD (2019) Cross-species hybridization and the origin of North African date palms. Proc Natl Acad Sci USA 116:1651–1658. https://doi.org/10.1073/pnas.1817453116
Food and Agriculture Organization of the United Nations (2019) http://www.fao.org/faostat/en/#data/QC
Gros-Balthazard M, Hazzouri KM, Flowers JM (2018) Genomic insights into date palm origins. Genes 9:502. https://doi.org/10.3390/genes9100502
Hammer Ø, Harper DA, Ryan PD (2001) PAST: Paleontological statistics software package for education and data analysis. Palaeontol Elect 4:9
Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM (2004) Identification of Birds through DNA Barcodes. PLoS Biol 2:e312. https://doi.org/10.1371/journal.pbio.0020312
Jain, S. M., & Johnson, D. V (2015) Date palm genetic resources and utilization. J. M. Al-Khayri (Ed.). Springer.
Jaradat AA, Zaid A (2004) Quality traits of date palm fruits in a center of origin and center of diversity. Food Agric Environ 2:208–217
Jurado-Rivera JA, Vogler AP, Reid CA, Petitpierre E, Gómez-Zurita J (2009) DNA barcoding insect-host plant associations. Proc Biol Sci 276:639–648. https://doi.org/10.1098/rspb.2008.1264
Koohdar F, Sheidai M (2019) Molecular investigation in few spices of Dacocephalum in Iran: Species relationship, reticulation and divergence time. Ind Crops Prod 141:111758. https://doi.org/10.1016/j.indcrop.2019.111758
Kress WJ, Erickson DL, Swenson NG, Thompson J, Uriarte M et al (2010) Advances in the use of DNA barcodes to build a community phylogeny for tropical trees in a puerto rican forest dynamics plot. PLoS ONE 5:e15409. https://doi.org/10.1371/journal.pone.0015409
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S (2015) Plant DNA barcoding: from gene to genome. Biol Rev 90:157–166
Marsafari M, Mehrabi AA (2013) Molecular identification and genetic diversity of iranian date palm (Phoenix dactylifera L.) cultivars using ISSR and RAPD markers. Aust J Crop Sci 7:1160
Mohamoud YA, Mathew LS, Torres MF et al (2019) Novel subpopulations in date palm (Phoenix dactylifera) identified by population-wide organellar genome sequencing. BMCGenom 20:498. https://doi.org/10.1186/s12864-019-5834-7
Naeem A, Khan AA, Cheema HMN, Khan IA, Buerkert A (2014) DNA barcoding for species identification in the Pal+mae family. Genet Mol Res 13:10341–10348. https://doi.org/10.4238/2014.December.4.29
Nemati Z, Zeinalabedini M, Majidian P, Eftekharian Jahromi A, Kiani D (2014) Phylogenetic relationships among Iranian and Spanish date palms (Phoenix dactylifera L.) revealed by microsatellite markers. J Hortic Sci Biotechnol 89:115–120. https://doi.org/10.1080/14620316.2014.11513056
Partovi R, Iranbakhsh A, Sheidai M, Ebadi M (2020) Population genetic studies in wild olive (Olea cuspidata) by molecular barcodes and SRAP molecular markers. Caryologia. https://doi.org/10.13128/caryologia-147
Saboori S, Noormohammadi Z, Sheidai M, Marashi S (2019) SCoT molecular markers and genetic fingerprinting of date palm (Phoenix dactylifera L.) cultivars. Genet Resour Crop Evol 67:73–82. https://doi.org/10.1007/s10722-019-00854-x
Saboori S, Noormohammadi Z, Sheidai M, Marashi S (2020) Insight into date palm diversity: genetic and morphological investigations. Plant Mol Biol Rep. https://doi.org/10.1007/s11105-020-01241-y
Shapcott A, Forster PI, Guymer GP, McDonald WJF, Faith DP, Erickson D et al (2015) Mapping biodiversity and setting conservation priorities for SE Queensland’s rainforests using DNA barcoding. PLoS ONE 10:e0122164. https://doi.org/10.1371/journal.pone.0122164
Sheidai M, Darini S, Talebi SM, Koohdar F, Ghasemzadeh-Baraki S (2019) Molecular systematic study in the genus Linum (Linaceae) in Iran. Acta Bot Hung 61:421–434. https://doi.org/10.1556/034.61.2019.3-4.11
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. https://paup.phylosolutions.com
Tabaripoor R, Sheidai M, Talebi SM, Noormohammadi Z (2016) Population genetic diversity and structure in Ziziphora tenuior L.: identification of potential gene pools. Genetika 48:565–578. https://doi.org/10.2298/GENSR1602565T
Zohary D, Hopf M (2000) Domestication of plants in the old world: the origin and spread of cultivated plants in West Asia, Europe and the Nile Valley (No. Ed. 3). Oxford University Press. ISBN:9780199549061
Acknowledgements
We acknowledge Science and Research Branch, Islamic Azad University for providing laboratory. We thank the Iran National Science Foundation (INSF), for partial financial support of this Project (No. 97010700).
Funding
None.
Author information
Authors and Affiliations
Contributions
Z.N. and M.Sh: conceptualization of the project; M.Sh.: analyses of data; S.S: data collection and lab work; S.M.: providing samples. The authors accept responsibility for releasing this material. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for Publication
The authors accept responsibility for releasing this material.
Supplementary Information
Below is the link to the Supplementary Information.
Rights and permissions
About this article
Cite this article
Saboori, S., Noormohammadi, Z., Sheidai, M. et al. Date Palm (Phoenix dactylifera L.) Cultivar Relationships Based on Chloroplast Genotyping. Iran J Sci Technol Trans Sci 45, 833–840 (2021). https://doi.org/10.1007/s40995-021-01085-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40995-021-01085-5