Abstract
Purpose of Review
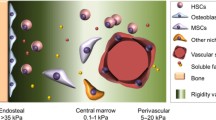
Adult human hematopoiesis resides in the bone marrow (BM), which is comprised of multiple niches capable of supporting hematopoietic stem and progenitor cells (HSPCs), as well as their downstream lineages. In maintaining the blood-forming cells of the body, the BM microenvironment is spatially regulated by a number of stromal support cells to maintain HSPCs and promote their differentiation in response to exogenous stimuli (e.g., injury, regeneration, disease). Although mouse models are the gold standard for studying hematopoietic biology, in vitro models are gaining acceptance in studies of human-centered, patient-specific functions of blood and immune progenitors.
Recent Findings
Over the past 5–10 years, model systems of the BM, including ossicles, engineered tissues, and organs-on-a-chip, have emerged as new vehicles to study both healthy and malignant hematopoiesis.
Summary
In this review, we describe progress in bioengineered tools for studying the BM, as well as design considerations necessary for developing more advanced systems for precision medicine and translational applications.




Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
Dzierzak E, Bigas A. Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell. 2018;22(5):639–51.
Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327–34.
Pinho S, Frenette PS. Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol. 2019;20(5):303–20.
Ranzoni AM, Tangherloni A, Berest I, et al. Integrative single-cell RNA-seq and ATAC-seq analysis of human developmental hematopoiesis. Cell Stem Cell. 2021;28(3):472-487.e7.
Bowman RL, Busque L, Levine RL. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell. 2018;22(2):157–70.
Shanks N, Greek R, Greek J. Are animal models predictive for humans? Philos Ethics Humanit Med. 2009;4:2.
Yin X, Benjamin, Mead E, et al. Engineering stem cell organoids. Cell Stem Cell. 2016;18(1):25–38.
Tavakol DN, Fleischer S, Vunjak-Novakovic G. Harnessing organs-on-a-chip to model tissue regeneration. Cell Stem Cell. 2021;28(6):993–1015.
Ronaldson-Bouchard K, Vunjak-Novakovic G. Organs-on-a-chip: a fast track for engineered human tissues in drug development. Cell Stem Cell. 2018;22(3):310–24.
Bourgine PE, Klein T, Paczulla AM, et al. In vitro biomimetic engineering of a human hematopoietic niche with functional properties. Proc Natl Acad Sci U S A. 2018;115(25):E5688-e5695.
Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76.
Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10(6):678–84.
Sharma A, Sances S, Workman MJ, et al. Multi-lineage human iPSC-derived platforms for disease modeling and drug discovery. Cell Stem Cell. 2020;26(3):309–29.
Hendriks D, Clevers H, Artegiani B. CRISPR-Cas tools and their application in genetic engineering of human stem cells and organoids. Cell Stem Cell. 2020;27(5):705–31.
Low LA, Mummery C, Berridge BR, et al. Organs-on-chips: into the next decade. Nat Rev Drug Discov. (2020).
Fleischer S, Tavakol DN, Vunjak-Novakovic G. From arteries to capillaries: approaches to engineering human vasculature. Adv Funct Mater. n/a(n/a) 2020;1910811.
Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132(4):598–611.
Rezza A, Sennett R, Rendl M. Chapter twelve - adult stem cell niches: cellular and molecular components, in: M. Rendl (Ed.), Current topics in developmental biology, Academic Press. 2014;333–372.
Yu VW, Scadden DT. Hematopoietic stem cell and its bone marrow niche. Curr Top Dev Biol. 2016;118:21–44.
Ramasamy SK, Kusumbe AP, Adams RH. Regulation of tissue morphogenesis by endothelial cell-derived signals. Trends Cell Biol. 2015;25(3):148–57.
Ding L, Saunders TL, Enikolopov G, et al. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481(7382):457–62.
Genet N, Bhatt N, Bourdieu A, et al. Multifaceted roles of connexin 43 in stem cell niches. Current Stem Cell Reports. 2018;4(1):1–12.
Winkler IG, Barbier V, Nowlan B, et al. Vascular niche E-selectin regulates hematopoietic stem cell dormancy, self renewal and chemoresistance. Nat Med. 2012;18(11):1651–7.
Asada N, Kunisaki Y, Pierce H, et al. Differential cytokine contributions of perivascular haematopoietic stem cell niches, Nat Cell Biol. 2017;19(3):214–223.
Kunisaki Y, Bruns I, Scheiermann C, et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502(7473):637–43.
Naveiras O, Nardi V, Wenzel PL, et al. Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature. 2009;460(7252):259–63.
Zhou BO, Yu H, Yue R, et al. Bone marrow adipocytes promote the regeneration of stem cells and haematopoiesis by secreting SCF. Nat Cell Biol. 2017;19(8):891–903.
Mansour A, Abou-Ezzi G, Sitnicka E, et al. Osteoclasts promote the formation of hematopoietic stem cell niches in the bone marrow. J Exp Med. 2012;209(3):537–49.
Taichman RS, Emerson SG. The role of osteoblasts in the hematopoietic microenvironment. Stem Cells. 1998;16(1):7–15.
Divieti Pajevic P, Krause DS. Osteocyte regulation of bone and blood. Bone 2019;119:13–18.
Maryanovich M, Takeishi S, Frenette PS. Neural regulation of bone and bone marrow. Cold Spring Harb Perspect Med 2018;8(9).
Mendez-Ferrer S, Lucas D, Battista M, et al. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452(7186):442–7.
Theocharis AD, Skandalis SS, Gialeli C, et al. Extracellular matrix structure. Adv Drug Deliv Rev. 2016;97:4–27.
Gattazzo F, Urciuolo A, Bonaldo P. Extracellular matrix: a dynamic microenvironment for stem cell niche. Biochim Biophys Acta. 2014;1840(8):2506–19.
Yu VWC, Scadden DT. Heterogeneity of the bone marrow niche. Curr Opin Hematol. 2016;23(4):331–8.
Coutu DL, Kokkaliaris KD, Kunz L, et al. Three-dimensional map of nonhematopoietic bone and bone-marrow cells and molecules. Nat Biotechnol. 2017;35(12):1202–10.
Aasebø E, Birkeland E, Selheim F, et al. The extracellular bone marrow microenvironment—a proteomic comparison of constitutive protein release by in vitro cultured osteoblasts and mesenchymal stem cells. Cancers. 2021;13(1):62.
Zöller M. CD44, hyaluronan, the hematopoietic stem cell, and leukemia-initiating cells. Front Immunol. 2015;6(235).
Chen X-D, Dusevich V, Feng JQ, et al. Extracellular matrix made by bone marrow cells facilitates expansion of marrow-derived mesenchymal progenitor cells and prevents their differentiation into osteoblasts. J Bone Miner Res. 2007;22(12):1943–56.
Yamashita M, Dellorusso PV, Olson OC, et al. Dysregulated haematopoietic stem cell behaviour in myeloid leukaemogenesis. Nat Rev Cancer. 2020;20(7):365–82.
Mitroulis I, Kalafati L, Bornhäuser M, et al. Regulation of the bone marrow niche by inflammation. Front Immunol. 2020;11:1540–1540.
Witkowski MT, Dolgalev I, Evensen NA, et al. Extensive remodeling of the immune microenvironment in B cell acute lymphoblastic leukemia. Cancer Cell. 2020;37(6):867-882.e12.
Kode A, Manavalan JS, Mosialou I, et al. Leukaemogenesis induced by an activating β-catenin mutation in osteoblasts. Nature. 2014;506(7487):240–4.
Leimkühler NB, Gleitz HFE, Ronghui L, et al. Heterogeneous bone-marrow stromal progenitors drive myelofibrosis via a druggable alarmin axis. Cell Stem Cell. 2021;28(4):637-652.e8.
Chramiec A, Vunjak-Novakovic G. Tissue engineered models of healthy and malignant human bone marrow. Adv Drug Deliv Rev. 2019;140:78–92.
Reinisch A, Thomas D, Corces MR, et al. A humanized bone marrow ossicle xenotransplantation model enables improved engraftment of healthy and leukemic human hematopoietic cells. Nat Med. 2016;22(7):812–21.
Abarrategi A, Mian SA, Passaro D, et al. Modeling the human bone marrow niche in mice: from host bone marrow engraftment to bioengineering approaches. J Exp Med. 2018;215(3):729–43.
Tavakol DN, Tratwal J, Bonini F, et al. Injectable, scalable 3D tissue-engineered model of marrow hematopoiesis. Biomaterials. 2019;119665.
•• Chou DB, Frismantas V, Milton Y, et al. On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology. Nat Biomed Eng. 2020;4(4):394–406. This work describes the first multicellular, spatially-organized model of a human BM-on-a-chip to study application areas in radiation toxicity, blood disorder modeling, and therapeutic drug testing." but it would not let us bold the descriptive statement.
Nelson MR, Ghoshal D, Mejías JC, et al. A multi-niche microvascularized human bone marrow (hBM) on-a-chip elucidates key roles of the endosteal niche in hBM physiology. Biomaterials. 2021;270:120683.
Dupard SJ, Grigoryan A, Farhat S, et al. Development of humanized ossicles: bridging the hematopoietic gap. Trends Mol Med. 2020;26(6):552–69.
Krupnick AS, Shaaban A, Radu A, et al. Bone marrow tissue engineering. Tissue Eng. 2002;8(1):145–55.
Shah NJ, Mao AS, Shih T-Y, et al. An injectable bone marrow–like scaffold enhances T cell immunity after hematopoietic stem cell transplantation. Nat Biotechnol. 2019;37(3):293–302.
Ferreira MS, Jahnen-Dechent W, Labude N, et al. Cord blood-hematopoietic stem cell expansion in 3D fibrin scaffolds with stromal support. Biomaterials. 2012;33(29):6987–97.
Leisten I, Kramann R, Ventura Ferreira MS, et al. 3D co-culture of hematopoietic stem and progenitor cells and mesenchymal stem cells in collagen scaffolds as a model of the hematopoietic niche. Biomaterials. 2012;33(6):1736–47.
Mahadik BP, Bharadwaj NAK, Ewoldt RH, et al. Regulating dynamic signaling between hematopoietic stem cells and niche cells via a hydrogel matrix. Biomaterials. 2017;125:54–64.
Gilchrist AE, Lee S, Hu Y, et al. Soluble signals and remodeling in a synthetic gelatin-based hematopoietic stem cell niche. Adv Healthcare Mater. 2019;8(20):1900751.
Gilchrist AE, Harley BAC. Connecting secretome to hematopoietic stem cell phenotype shifts in an engineered bone marrow niche. Integr Biol (Camb). 2020;12(7):175–87.
Torisawa Y-S, Spina CS, Mammoto T, et al. Bone marrow–on–a–chip replicates hematopoietic niche physiology in vitro. Nat Methods. 2014;11(6):663–9.
Glaser DE, Curtis MB, Sariano PA, et al. Organ-on-a-chip model of vascularized human bone marrow niches, bioRxiv 2020.04.17.039339.
Aleman J, George SK, Herberg S, et al. Deconstructed microfluidic bone marrow on-a-chip to study normal and malignant hemopoietic cell–niche interactions. Small. 2019;15(43):1902971.
Belloni D, Heltai S, Ponzoni M, et al. Modeling multiple myeloma-bone marrow interactions and response to drugs in a 3D surrogate microenvironment. Haematologica. 2018;103(4):707–16.
Ma C, Witkowski MT, Harris J, et al. Leukemia-on-a-chip: dissecting the chemoresistance mechanisms in B cell acute lymphoblastic leukemia bone marrow niche, Science Advances 2020;6(44):eaba5536.
Chramiec A, Teles D, Yeager K, et al. Integrated human organ-on-a-chip model for predictive studies of anti-tumor drug efficacy and cardiac safety. Lab Chip. 2020;20(23):4357–72.
Sieber S, Wirth L, Cavak N, et al. Bone marrow-on-a-chip: long-term culture of human haematopoietic stem cells in a three-dimensional microfluidic environment. J Tissue Eng Regen Med. 2018;12(2):479–89.
Marturano-Kruik A, Nava MM, Yeager K, et al. Human bone perivascular niche-on-a-chip for studying metastatic colonization. Proc Natl Acad Sci U S A. 2018;115(6):1256–61.
Congrains A, Bianco J, Rosa RG, et al. 3D scaffolds to model the hematopoietic stem cell niche: applications and perspectives, Materials (Basel) 2021;14(3).
Moore CA, Siddiqui Z, Carney GJ, et al. A 3D bioprinted material that recapitulates the perivascular bone marrow structure for sustained hematopoietic and cancer models, Polymers (Basel) 2021;13(4).
Zhou D, Chen L, Ding J, et al. A 3D engineered scaffold for hematopoietic progenitor/stem cell co-culture in vitro. Sci Rep. 2020;10(1):11485.
Sugiyama T, Kohara H, Noda M, et al. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25(6):977–88.
Kunz L, Schroeder T. A 3D tissue-wide digital imaging pipeline for quantitation of secreted molecules shows absence of CXCL12 gradients in bone marrow. Cell Stem Cell. 2019;25(6):846-854.e4.
Christodoulou C, Spencer JA, Yeh S-CA, et al. Live-animal imaging of native haematopoietic stem and progenitor cells. Nature. 2020;578(7794):278–83.
Tratwal J, Bekri D, Boussema C, et al. MarrowQuant across aging and aplasia: a digital pathology workflow for quantification of bone marrow compartments in histological sections, Frontiers in Endocrinology 2020;11(480).
Haas S. Hematopoietic stem cells in health and disease—insights from single-cell multi-omic approaches. Current Stem Cell Reports. 2020;6(3):67–76.
Baccin C, Al-Sabah J, Velten L, et al. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat Cell Biol. 2020;22(1):38–48.
Graney PL, Tavakol DN, Chramiec A, et al. Engineered models of tumor metastasis with immune cell contributions. iScience 2021;102179.
Sugimura R, Jha DK, Han A, et al. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature. 2017;545(7655):432–8.
Sturgeon CM, Ditadi A, Awong G, et al. Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat Biotechnol. 2014;32(6):554–61.
Sandler VM, Lis R, Liu Y, et al. Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature. 2014;511(7509):312–8.
Kotini AG, Chang C-J, Chow A, et al. Stage-specific human induced pluripotent stem cells map the progression of myeloid transformation to transplantable leukemia. Cell Stem Cell. 2017;20(3):315-328.e7.
Doulatov S, Vo LT, Macari ER, et al. Drug discovery for Diamond-Blackfan anemia using reprogrammed hematopoietic progenitors, Science Translational Medicine 2017;9(376):eaah5645.
Wang T, Pine AR, Kotini AG, et al. Sequential CRISPR gene editing in human iPSCs charts the clonal evolution of myeloid leukemia and identifies early disease targets, Cell Stem Cell (2021). This study demonstrated an expansion on previous iPSC-derived hematopoietic stem cell protocols, identifying stage-specific differences in modeling hematological malignancies in vitro.
Acknowledgements
Figures were created in part with Biorender.com.
Funding
This study was financially supported by NIH (grants EB025765, EB027062, CA249799), NSF (Graduate Research Fellowship DGE1644869), NYSTEM (grant C32606GG), NASA (RAD0104), TRISH (NNX16A069A), Columbia 2020 SEAS-HICCC Pilot Grant, and Blavatnik Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Stem Cell Switches and Regulators
Rights and permissions
About this article
Cite this article
Tavakol, D.N., Chen, J., Chavkin, N.W. et al. Lessons from Biology: Engineering Design Considerations for Modeling Human Hematopoiesis. Curr Stem Cell Rep 7, 174–184 (2021). https://doi.org/10.1007/s40778-021-00195-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40778-021-00195-5